Difference between revisions of "Database"
From Berlin Harvesting and Indexing Toolkit
| (One intermediate revision by the same user not shown) | |||
| Line 10: | Line 10: | ||
*the bio_datasource table, which stores every datasource (accespoint, name, number of records, standard and protocol). | *the bio_datasource table, which stores every datasource (accespoint, name, number of records, standard and protocol). | ||
*the tripleidstore table, which stores every triple ID (unitID, collectionCode, institutionCode) met during the processing of the harvested records. | *the tripleidstore table, which stores every triple ID (unitID, collectionCode, institutionCode) met during the processing of the harvested records. | ||
| + | |||
| + | [[File:rawtables.png|800px]] | ||
| + | <BR> | ||
Latest revision as of 15:45, 16 November 2015
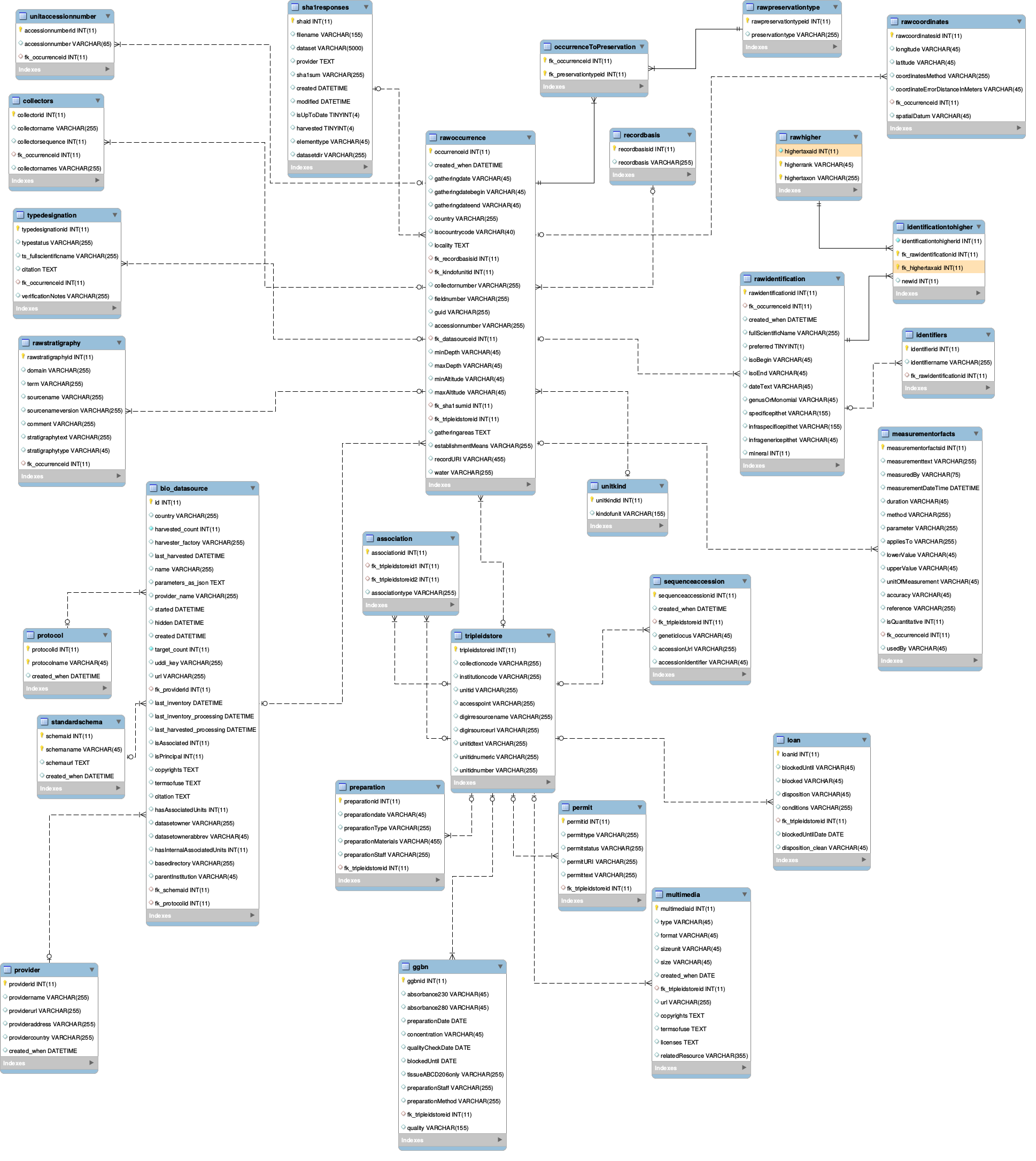
The default database is MySQL. If you want to use another system, you will have to change the configuration (application.properties) and add the library.
Data is saved two times:
- first, the original data delivered by the provider is saved in the raw* tables (rawidentification, rawcoordinates, rawoccurrence, rawpreservationtype, rawhigher)
- after running the quality tests, improved data is saved in tables without the raw prefix (identification, coordinates, occurrence, perservationtype, higher)
There are 2 central tables:
- the bio_datasource table, which stores every datasource (accespoint, name, number of records, standard and protocol).
- the tripleidstore table, which stores every triple ID (unitID, collectionCode, institutionCode) met during the processing of the harvested records.