Installation
Berlin Harvesting and Indexing Toolkit
Pre-requisites
Basics
Tested with Java JVM >=1.8, on Linux and Windows (might not start with JVM 1.7 and older).
Can be deployed with Tomcat (tested with Apache Tomcat/6.0.39 JRE 1.8.0_05-b13) or Jetty (tested with Jetty 8.1.14), or can be run inside Eclipse (tested with Eclipse Luna and Kepler. If the libraries get deleted by eclipse, run mvn eclipse:eclipse to get automatically all libraries defined in the pom-xml file ; project encoding must be UTF-8 (also defined in the pom.xml)). Please download the war file from [1].
MySQL Database v5.5
Required web-services for data quality checks
For data quality checks related to geography B-HIT uses gisgraphy and CoordinatesKdTree. We recommend a local installation for both.
Local gisgraphy installation (coordinates check) or maybe online version, see http://www.gisgraphy.com/free-access.htm
CoordinatesKDTree werbservice (based on https://github.com/AReallyGoodName/OfflineReverseGeocode ). It can be found in the SVN http://ww2.biocase.org/svn/binhum/trunk/BinHum/Webservice/CoordinatesWS/, also as a WAR file available. It's a large WAR file, so you might have to change the Tomcat configuration off your manager (org.apache.tomcat.util.http.fileupload.FileUploadBase$SizeLimitExceededException) in order to allow it to be deployed (for example see https://www.netroby.com/view/3585).
Configuration
Before starting the app
Run the database creation script (schemaOnly.sql) and prefill the database with schemaProtocol.sql (contains default biodiversity schema names and protocols). Both files can be found in the root folder of the SVN.
Edit the application.properties file
The file is located in binhum/WebContent/WEB-INF/classes. Note:you can edit the file in the WAR file or if you already deployed it, edit the file in your tomcat webapp folder (if so, stop tomcat while editing).
| Quality tests
| |||||||
| A local folder for temporary file manipulation
Windows paths: please use the format C:\\Users\\username\\folder\\ |
temporaryFolder=/home/user/test | ||||||
| Set the installation type (production true/false) - if set to false, units will not be redownloaded if the files already exist on the disc. Should always be true in production or in doubt. | production=true | ||||||
| Database properties
| |||||||
| Application properties
| |||||||
Edit applicationContext-security.xml
The file is located in binhum/WebContent/WEB-INF/. Edit the admin password, md5 encoded. The default password corresponds to « banana! »
<user name="admin" password="bb7a307e32b93a931da89d0a214dd47f" authorities="ROLE_ADMIN" />
Be sure your DB schema is up to date
Check the file https://git.bgbm.org/ggbn/bhit/-/blob/master/BinHum/Harvester/sqlChanges.sql for the last modifications in the database schema! The best solution is to run each line separately, MySQL might complain that the column does already exist if you have just configured the whole thing for the first time, as the changes might already be in the schemaOnly.sql file.
Starting B-HIT
Based on your configuration, and the name chosen (eclipse config / war file name), open your favorite web-browser to http://localhost:8040/Bibhum/datasource/list.html . If you get a JSP error, check if there is any servlet-api*.jar in the B-HIT WEB-INF/lib/ Folder - is yes, delete it.
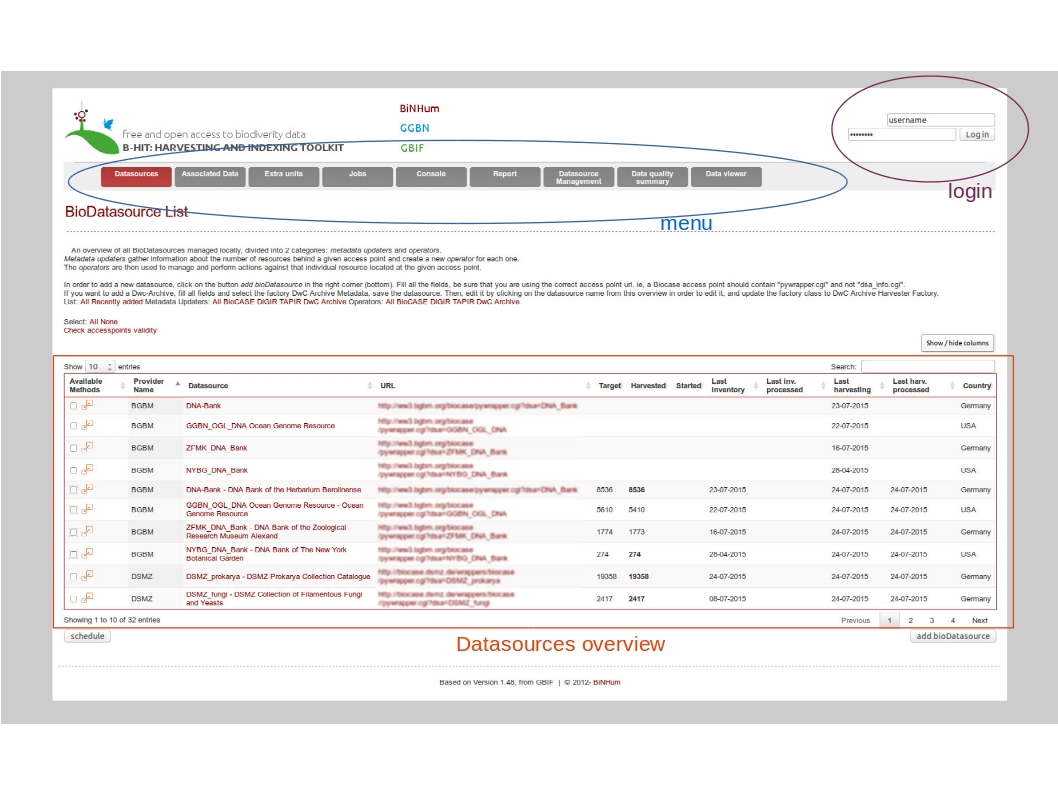
Overview
Main menu:
- Datasources : main entry point to add datasources/datasets, launch metadata operations, inventory+harvesting+processing data
- Associated datasources: entry point for associated data (relationships), to harvest and process associated data only
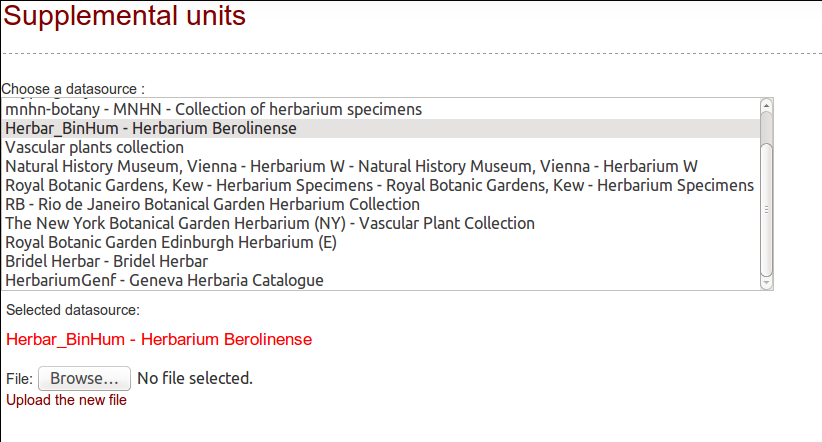
- Extra units: entry point for single units retrieval, based on a list of unit IDs
- Jobs: overview of waiting and running jobs
- Console: overview of log events
- Report: generation of reports (statistics)
- Datasource management: to hide or delete datasets
- Data quality : to launch quality tests, exports test results and display results
- Data viewer: to display data stored in the database, either the raw data or the improved data from the quality tests
Add a datasource
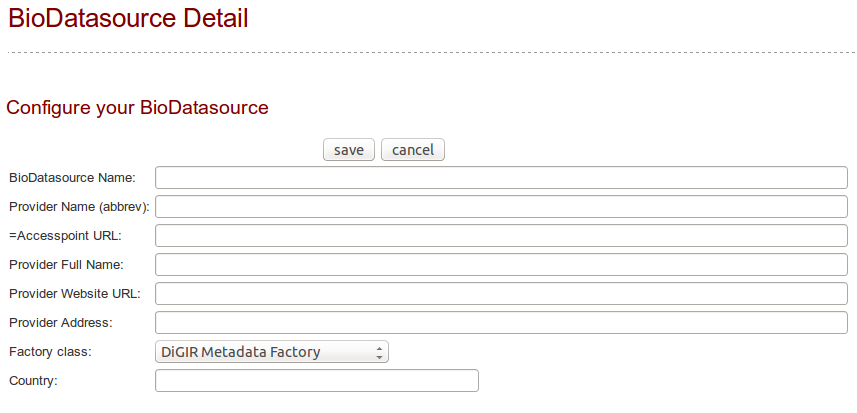
Click on the button « add bioDatasource » (bottom right). A form is displayed with mandatory fields to populate.
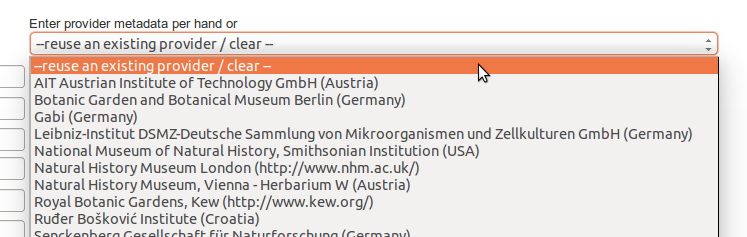
Enter a name for the datasource, the provider name abbreviated (ie. BGBM), the provider fullname (ie. Botanischer Garten und Botanisches Museum Berlin-Dahlem), the accesspoint (ie. http://ww3.bgbm.org/biocase/pywrapper.cgi?dsa=BoBo), the provider website (ie. http://www.bgbm.org), the provider postal address and country, and the type of datasource (ie. Biocase/digir/Tapir/darwincore archive). Then save. The new datasource is now listed on the http://localhost:8040/Bibhum/datasource/list.html page.
If you already added some providers, you can reuse them by selecting an existing provider from the dropdown menu on the right. It will populate automatically the form fields.
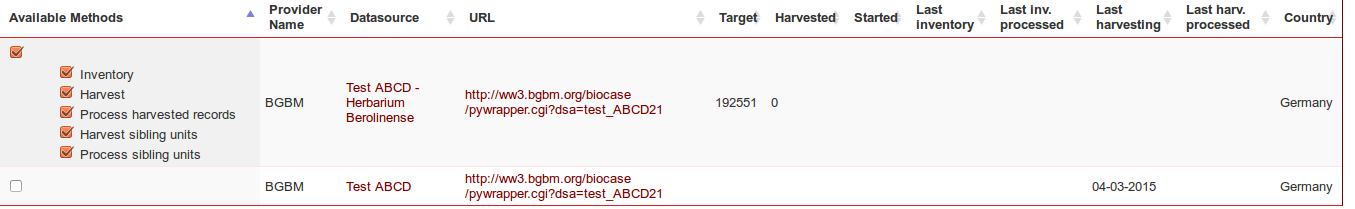
To see the available operations for this datasource (see Figure 4), click on the « available methods » checkbox on the left or on the expand/collapse icon.
Edit a datasource
To change the accesspoint or the name or the harvester type or the country, click on the datasource name. It will prompt a page where you can edit the details.
If you want to edit the schema (for example it selected ABCD 2.1 automatically, but you prefer to use ABCD 2.06), you will have to edit the database directly. You will have to change the value of bio_datasource.fk_schemaid (get the key from the standardschema table). If you switch from/to ABCD-Archive/ABCD, changing the harvester type will no be enough: you will also have to edit the bio_datasource.parameter_as_json column and add/edit a value for "abcdArchiveUrl".
Check accesspoints
On the datasource page, you can use the function “Check accesspoints validity” to check if the accesspoints are valid and accessible.
DEBUGGING HINT: The most common reason that a metadata update fails, is that the accesspoint is wrong. Classic Biocase URL looks like http://host/biocase/pywrapper.cgi?dsa=datasourceName
Retrieve datasets
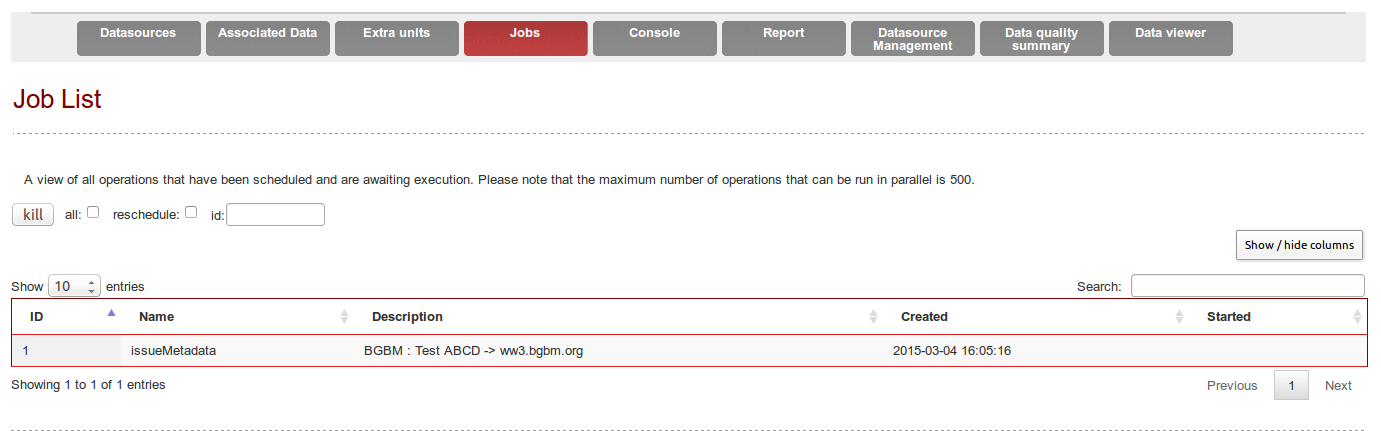
In order to trigger the « Metadata update » operation, check the box and click on « Schedule » at the bottom of the page.
A new Job is created and can be followed under the « Job » tab (/Bibhum/job/list.html).
Refresh the page to update the job status
Going back to the main page, a new line appeared in the list of datasource for the discovered dataset, with its corresponding available methods. These methods depend on the type of provider. ABCD-Archive will enable the methods Download ABCD-Archive/Process harvested, DwC-A will enable Download /Process harvested records
The target columns contains the theoretical number of units, according to the providers metadata.
DEBUGGING HINT: If the job is listed, and then disappears but nothing happened (reload the datasource page first), it might be due to a difference between the time stamp from the database server and the server where b-hit is running. B-HIT can only handle cases where the database server is 5min ahead from the application server.
Note: the datasource you entered manually will be a Metadata-Datasource. It will have the datasource name you typed. After the metadata operation has run, a second line will appear, very similar (same provider, same accesspoint), but the name might be different, as it will take the value returned by the datasource "itself".
Perform inventory
Only available for classic BIOCASe (and DiGIR) (not needed for ABCD-Archives or DwC A).
Select the first operation and click on « schedule ». The queries are stored on the disc, in compressed files. The location path is defined in the configuration file, and the directory for each datasource can be found in the MySQL table bio_datasource, column « basedirectory ».
Harvest data
Data will be harvested if and only if a list of units has been retrieved with the Inventory or can be done directly if the datasource provides an archive (ABCD or DwC). The outputs from this operation are * for classic datasources:
- One or more search requests (with enumerated extensions corresponding to the order in which they were dispatched, i.e. search_request.000)
- One or more search responses (with enumerated extensions corresponding to the order in which they were dispatched, i.e. search_response.000). Often, there will only be a single response per request, but sometimes there can be multiple responses for a single request!)
The default range is 200 units per request.* For archive datasources (ABCD-Archives, DwC-Archive)
- One search response, the downloaded archive unzipped in a folder named “archive”.
Process harvested records
In this operation, an Operator (BioDatasource) will collect all the search responses, parse them, and write the parsed values to the database. In case of re-harvesting a dataset, the system will check if the data changed since the last harvesting. It will calculate the new checksum of each downloaded file, and will modify data in the DB if and only if the checksum changed. Each checksum is stored in the DB table « sha1responses ». (See the extra doc file for more infos).
Tables starting with “raw” will be first filled (i.e. rawoccurrence, rawcoordinates).
Sibling units
Sibling units are units associated within a dataset (i.e split Herbarium sheets).
Associated datasets
Units can have associations to other specimen or observation data, within the same dataset or with an external dataset. These associations are stored in the association table (link between 2 units).
If it is required to add an external or new dataset, the datasource checkbox will be slightly different:
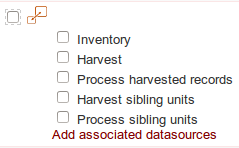
|
vs. | 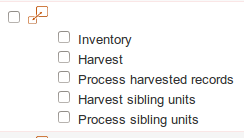
|
Display of dataset with or without associated datasources
Extra units
If you want to download only a subset of units, and you know their IDs (UnitID for ABCD or CatalogNumber for DarwinCore), you can do it!
First, create the datasource and retrieve its metatada.
Then, build a text file with the list of identifiers to get. One ID per line.
From the Extra Units tab, select the datasource you are interested in: a new line appears, you can now upload your text file.
If your datasource retrieves an ABCD Archive or a DarwinCore Archive, the complete datasource will be downloaded but only the IDs listed in the extra-file will be processed and save in the database.
Logs
Logs can be viewed in the Console tab. The Tomcat logs (ie. catalina.out can be helpful for debugging.
Datasource management
Datasources (harvester and metadata factories) can be deleted from the database with a simple button click. The selected source will be totally removed from the dabatase with all belonging records! Be careful!
You can also hide a datasource: a tag will be set in the database, and the data will be kept. The datasource will not be listed anymore in the datasource tab.
Quality
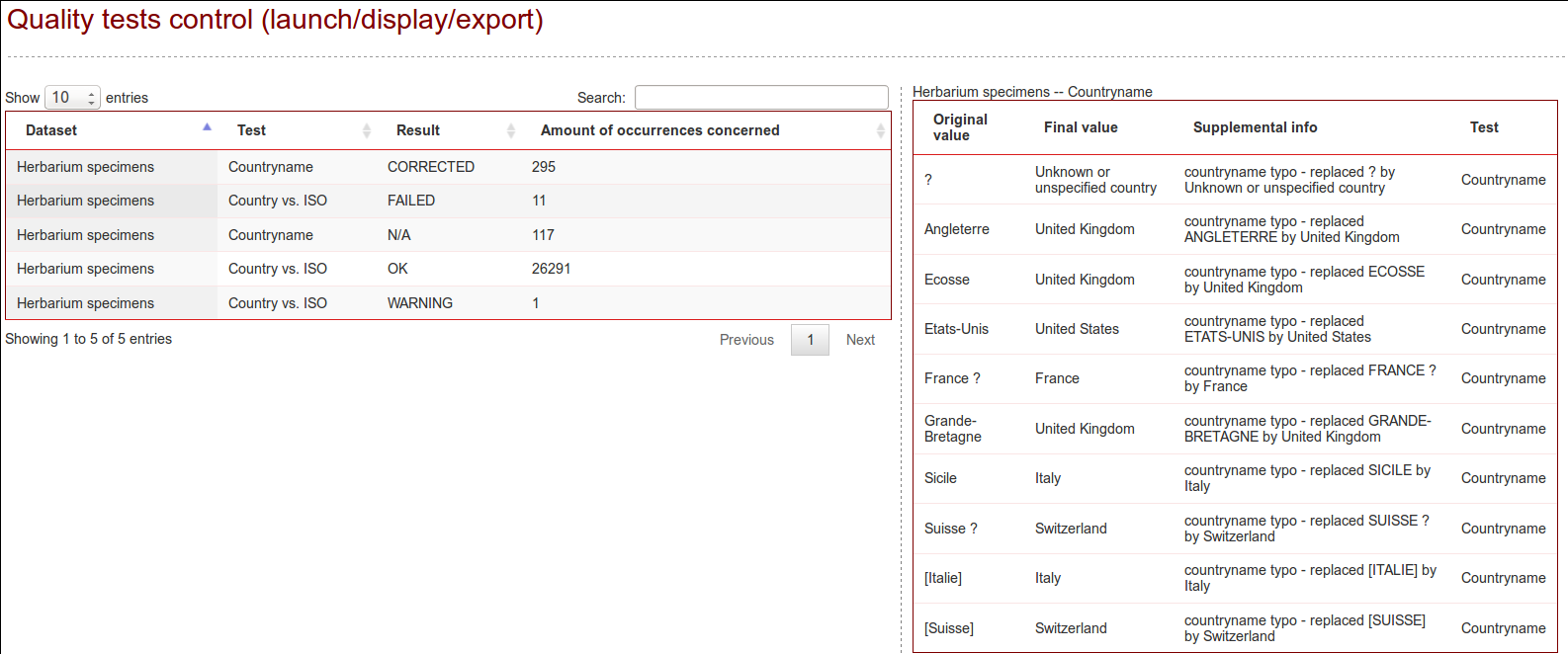
Quality tests can be triggered from the Quality tab if the option was activated (see configuration “qualityOnOff”).
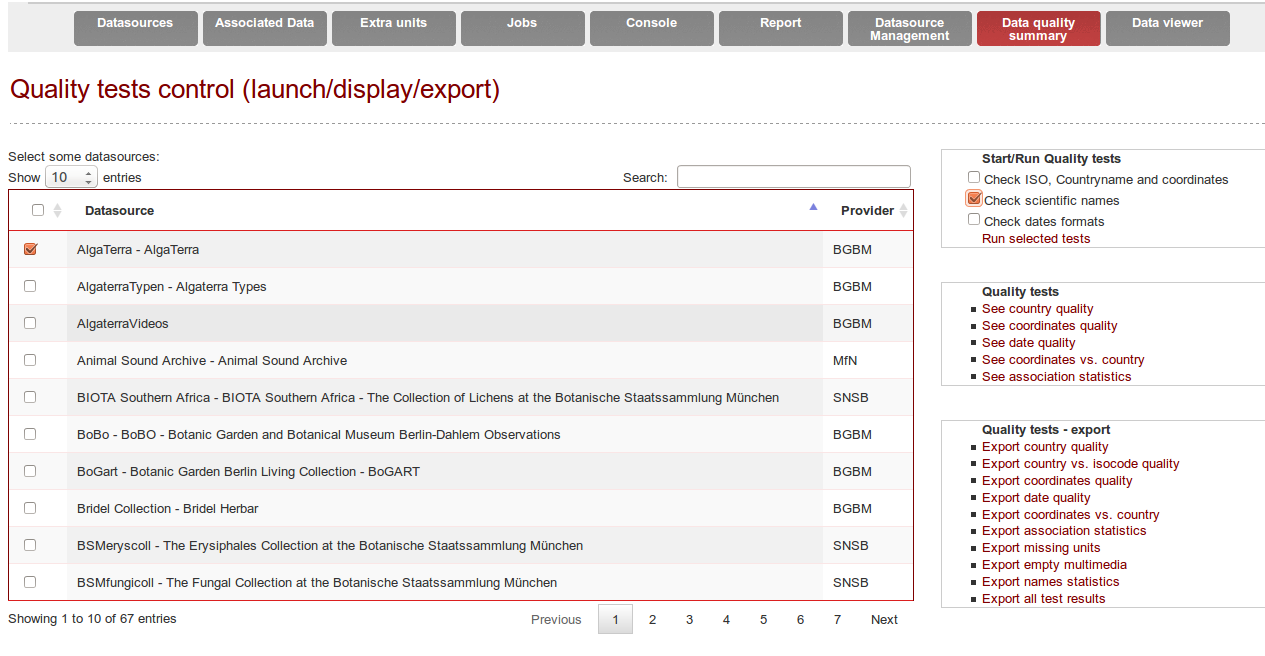
An extra column displays whether or not the quality tests have already been performed for each datasource.
Country names
The first test consists in translating the country name in English. The different accepted input values are extracted from the multiple language files and are completed based on the most common errors and typos met (i.e. “Iatlien” instead of “Italien”, missing empty spaces etc.). States and regions are also commonly mapped as countries by the providers: their affiliation has also been added to the known inputs
If the original country does no longer exists or if it borders do not fit any actual country, the corrected value will be set as an “Unknown or unspecified continent” (continent being replaced by Europe, Eurasia, Asia, North and Central America, South America, Oceania or Antarctica). “Unknown or unspecified country” will be inserted in the database for the empty values or for the characterisable values.
The second test consists in trying to extract the country based on the locality and the gathering areas.
It uses XML files to do the comparison between "known errors and typos" and isocodes. You can edit the files located in resources/org/binhum/quality/: alternative_country_names_v2.xml, countries_names.xml, countries_names_unambiguous.xml, oceans.xml, special_names.xml and water_country.xml if you want to correct or add something. Also have a look at the database tables country, countryToOceanOrContinent, and oceanOrContinent.
ISO-codes
The third test consists in standardising the ISO code in its 2-letter standard. The original value can be correct or non-available, can be replaced (ISO no more existing, i.e. SU) or corrected (3-letter code to 2-letter code).
The forth test consists in comparing the validated ISO and the validated country name. During this comparison, if no coordinates are available, a missing value will be inferred from the available country or from the available ISO (i.e. ISO-code=ZZ (unknown) and country name=Germany, coordinates empty-> ISO-code inferred as “DE”). Inferring a value will lead to a warning information in the database.
Coordinates
The fifth test will check the coordinates validity – the text values are parsed into decimal values, their ranges are checked (i.e. is the latitude between -90 and +90, resp. -180 and + 180 for the longitude).
The sixth test will compare the cleaned country data and the coordinates, using different methods and services. If the system previously detected an inconsistency between the ISO-code and the country name, it will try to improve the data based on the coordinates, and correct either the ISO-code or the country name.
If the original values of the coordinates do not fit neither the country field nor the ISO-code field, the quality process will check the opposites values of the coordinates (+latitude +longitude; +latitude -longitude; -latitude +longitude; -latitude -longitude), and try to add a leading number (many latitude were truncated and missed the first digit). It will also proceed to a permutation of the latitude and longitude values.
Tests on coordinates reuse existing code from Gisgraphy, KDTree and Geonames.
Date
Eventually, the gathering dates are checked and converted into the YYY-MM-DD format, and the gathering year is extracted.
Scientific names
During the quality task, the scientific names are parsed using the GBIF-Name Parser. Sadly, this tool cannot handle all names, logically the names containing errors, but also even some names which are correct according to taxonomic nomenclatures (emendavit, nominate…). About 50 regular expressions has been defined in order to parse the scientific names the GBIF-name-parser could not handle (Unicode characters, ex., forma, emend., names not conform to the different taxonomic nomenclatures, most common typos).
See the extra doc file for more infos regarding the quality tests.
Quality display and reports
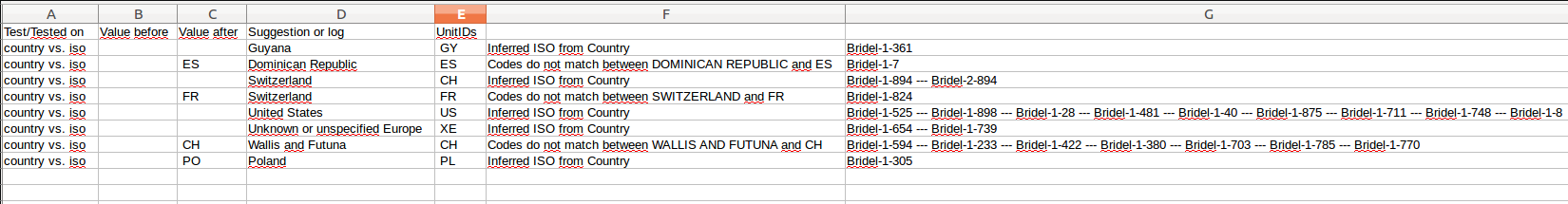
Every single quality test generates logs, which are saved in the database in specific tables.
From the Quality tab, it is also possible to extract these logs from the database and export them into text files with tab separated values. It will create one file per dataset and per test, if and only if the test modified the original content.
In worst case scenario, it will generate n x m files (n=number of datasources, m= number of tests run).
The system will only extract problematic rows, i.e. when the test failed or when it generated a warning.
This log files contain the name of the test executed, the original values, the edited values, and the list of units concerned by the changes.
These files can be sent to the data provider, who will then decide if they want to curate the data.
By clicking on Quality tests - export -> Export *, a new Job will be started. The quality file will be written on the disk where you configured the harvest.directory (in application.properties)
The quality results can also be displayed within B-HIT, by selecting a datasource and clicking on Quality tests -> See country quality
DEBUGGING HINT: If the job is listed, and then disappears but nothing happened (reload the datasource page first), it might be due to a difference between the time stamp from the database server and the server where b-hit is running. B-HIT can only handle cases where the database server is 5min ahead from the application server.
Reports
The lists of missing units (based on the datasources inventory files) can be generated from the report tab.
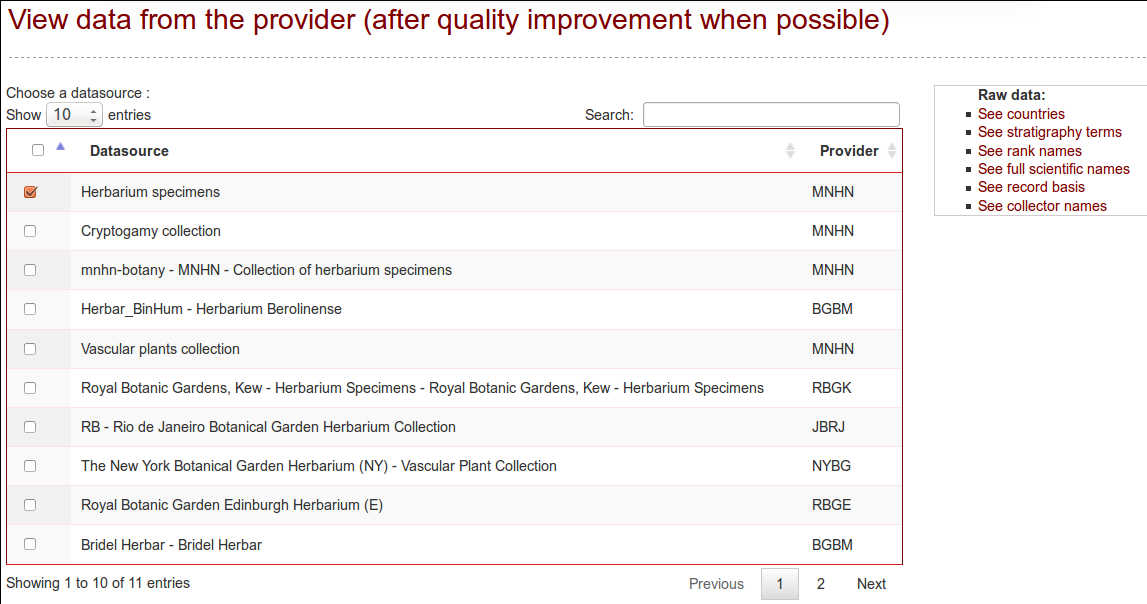
Data viewer
The cleaned data can be preview in this tab, for each datasource, only if the quality tests did run for that datasource.
Debug - where to go, what to do when something does not work as expected
It does not start
You are using Tomcat
If the app does not start after deployment ("The method getJspApplicationContext(ServletContext) is undefined for the type JspFactory"), remove servlet-api-2.3.jar from the webapp library folder (i.e. /var/lib/tomcat7/webapps/bhit/WEB-INF/lib/).
Database/MySQL error?
- Did you also update your DB schema as you updated B-HIT? Check the file https://git.bgbm.org/ggbn/bhit/-/blob/master/BinHum/Harvester/sqlChanges.sql.
Over suggestions
- Have a look in the log files from Tomcat in catalina.out or localhost_access_*.log
- Check where the problem is: is the provider responding? Did you check using the http://wiki.bgbm.org/bhit/index.php/Installation#Check_accesspoints functionality?
Are the responses valid? Check the .gz files saved in the harvest dir (cf. the application.properties file). The name of the directory for each datasource is in the bio_datasource table.