ER diagram: Difference between revisions
| Line 87: | Line 87: | ||
===Customer requests=== | ===Customer requests=== | ||
[[File:DNA Module Package7 Customer requests.jpg|thumb|200px|ER diagram package “Sequence data”]] | |||
This package includes management of all customer requests respectively orders. Since way of administer payments depends on institution this package is for the lab site only. | |||
*Main table: | |||
**request | |||
*Related packages: | |||
**Location stock/aliquots | |||
**DNA extractions | |||
===Specimen Tool=== | ===Specimen Tool=== | ||
===DNA Bank Network=== | ===DNA Bank Network=== | ||
Revision as of 15:14, 19 July 2011
Entity-relation diagram of the DNA Module V2.0
Overview
Bild einbauen
DNA extractions
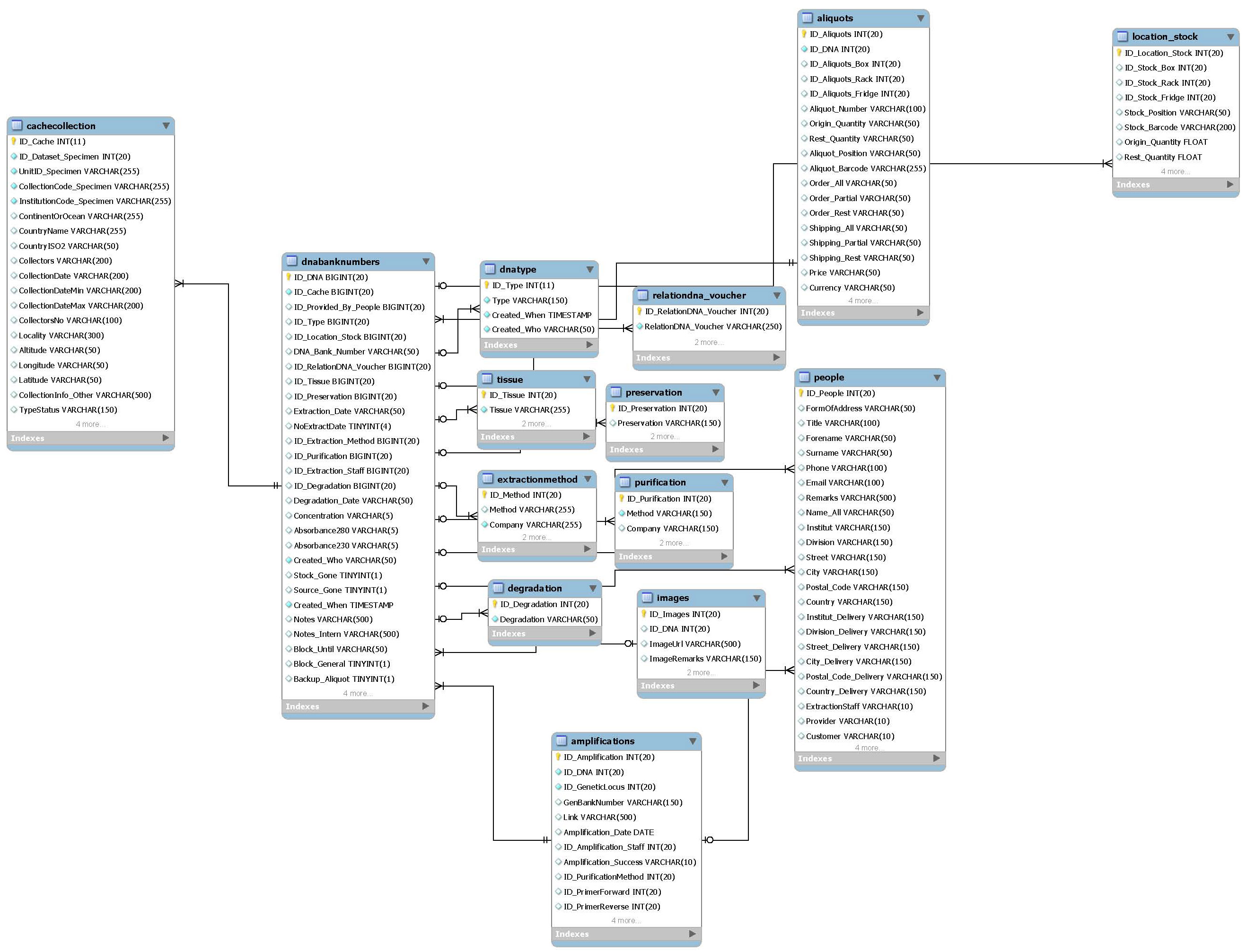
Main package of the DNA Module, related to many other packages. The main table is dnabanknumbers with several related list tables. The table people is connected to the dna extractions via three IDs: ExtractionStaff, Provided_By and AmplificationStaff.
- Main tables:
- dnabanknumbers
- people
- List tables:
- dnatype
- relationdna_voucher
- tissue
- preservation
- extractionmethod
- purification
- degradation
- images
- Related packages:
- Amplifications
- Locations Aliquots/stock
- Specimen cache
- Requests
- Molecular publications
- Log tables
Specimen data providers
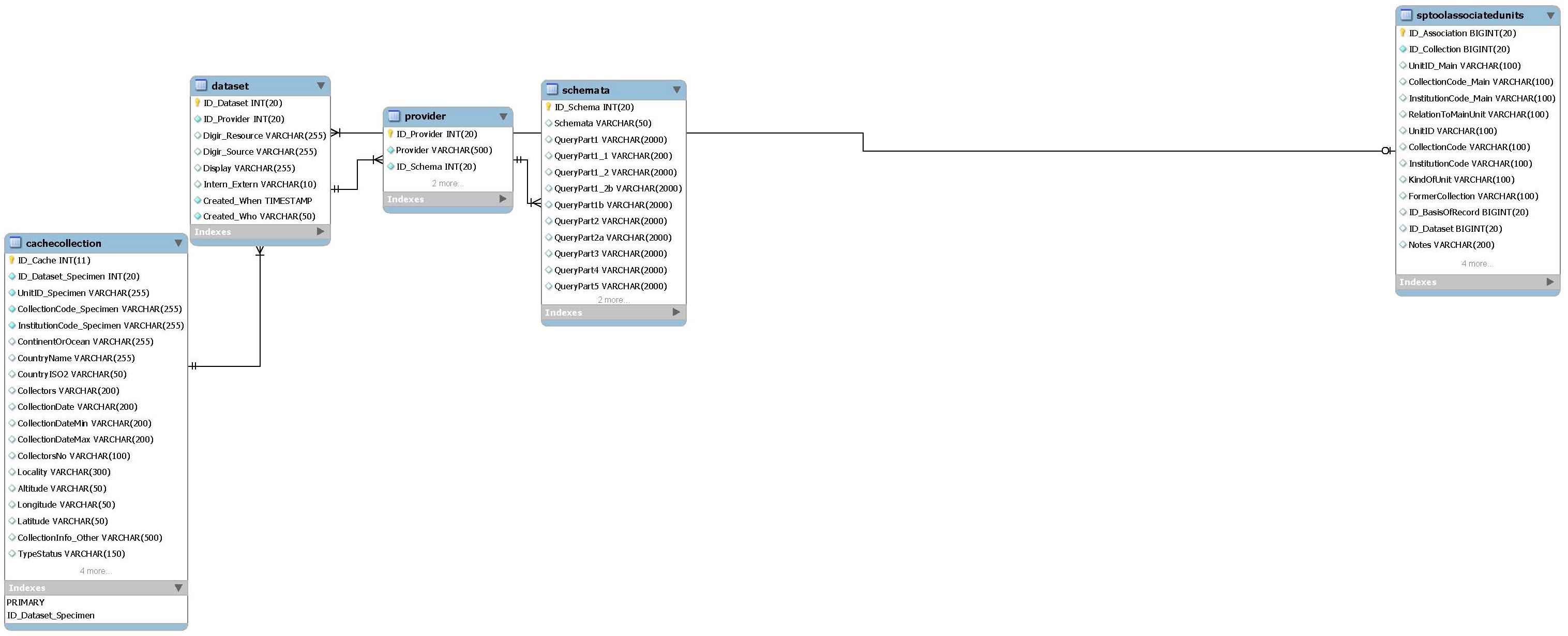
List of all connected specimen data providers (GBIF compliant databases). More than one dataset per provider is possible. The required schema and provider software is recorded by ID_Schema. The Specimen Tool is predefined with ID_Dataset = '1'. The Wrapper urls of both the Specimen Tool and the DNA data are left empty until definition via the Configuration Tool.
- Main table:
- dataset
- Related packages:
- Specimen cache
- Specimen Tool
Specimen cache
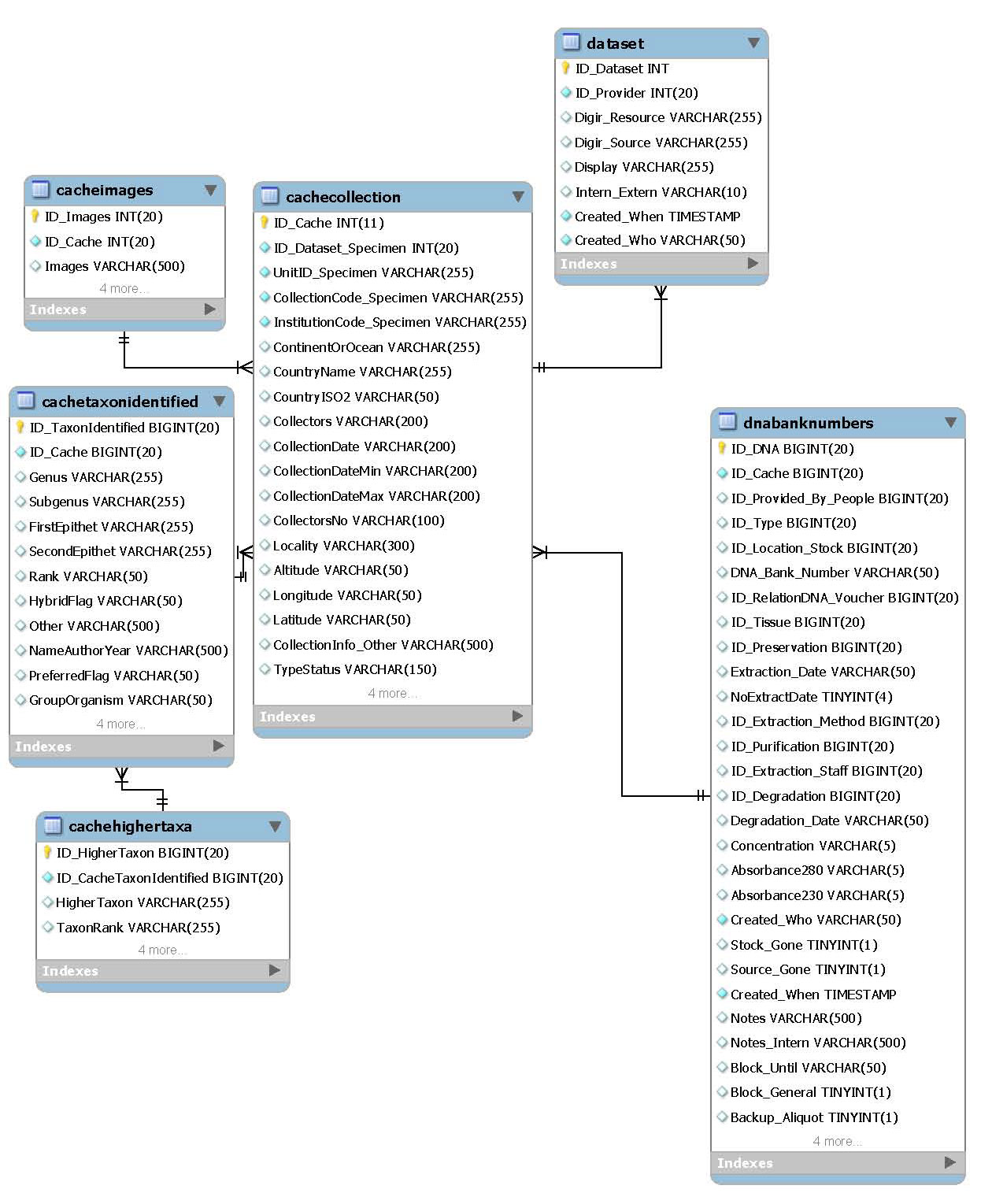
A few specimen parameters have to be recorded within the DNA Module database to enable search for exisiting taxa in the DNA database et cetera. The specimen data are recorded in four tables to get all available determinations and higher taxa. One specimen can be connected to several DNA extractions.
- Main table:
- cachecollection
- Related packages:
- Specimen data providers
- DNA extractions
Locations stock/aliquots
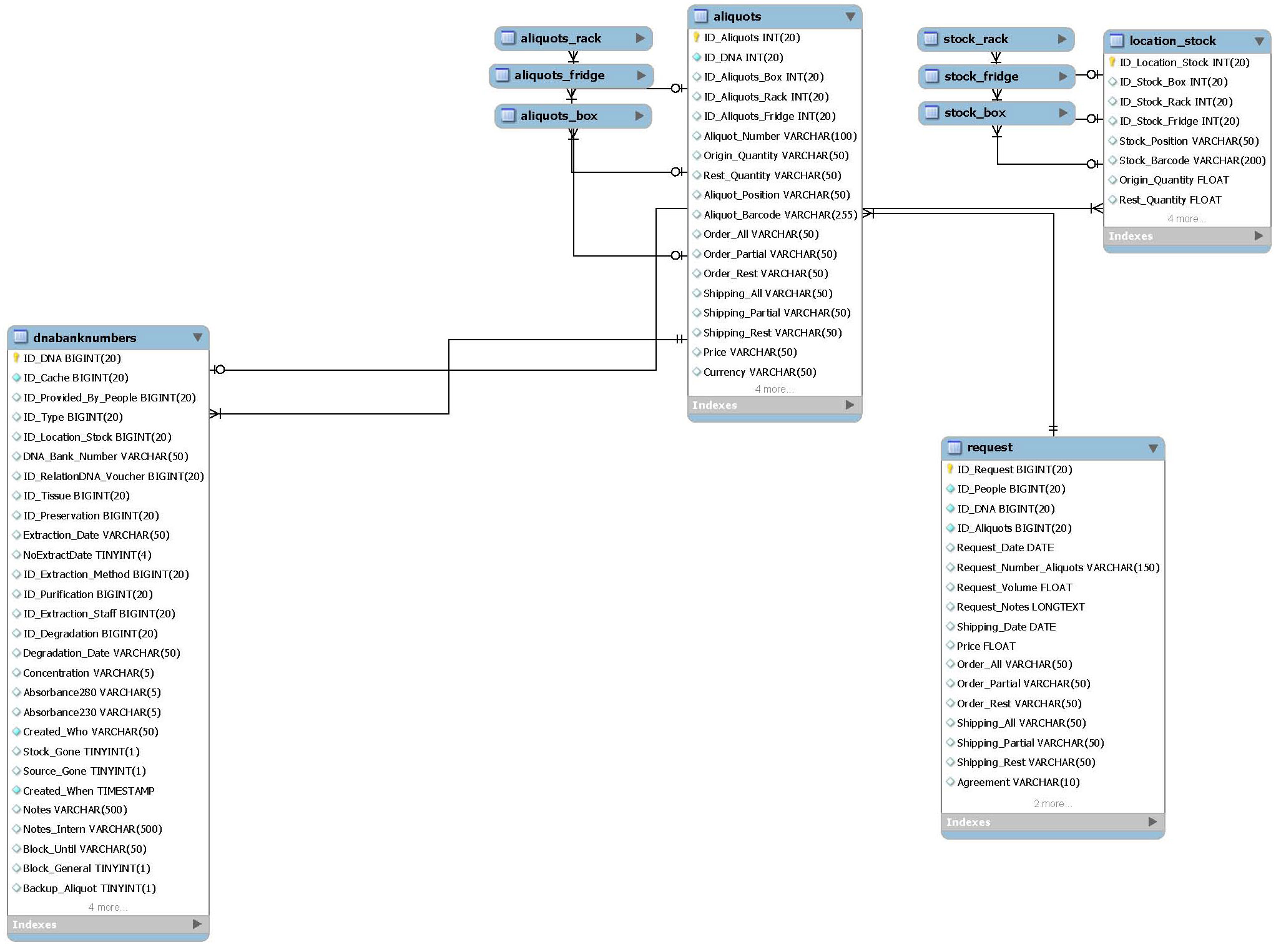
This package records the location and volumes of all aliquot and stock samples in the freezers. The information in this package is completely internal and help to find out easily which samples can be send to customers and which have to be extracted.
- Main tables:
- aliquots
- location_stock
- List tables:
- aliquots_box
- aliquots_fridge
- aliquots_rack
- stock_box
- stock_fridge
- stock_rack
- Related packages:
- DNA extraction
- Customer requests
Molecular publications
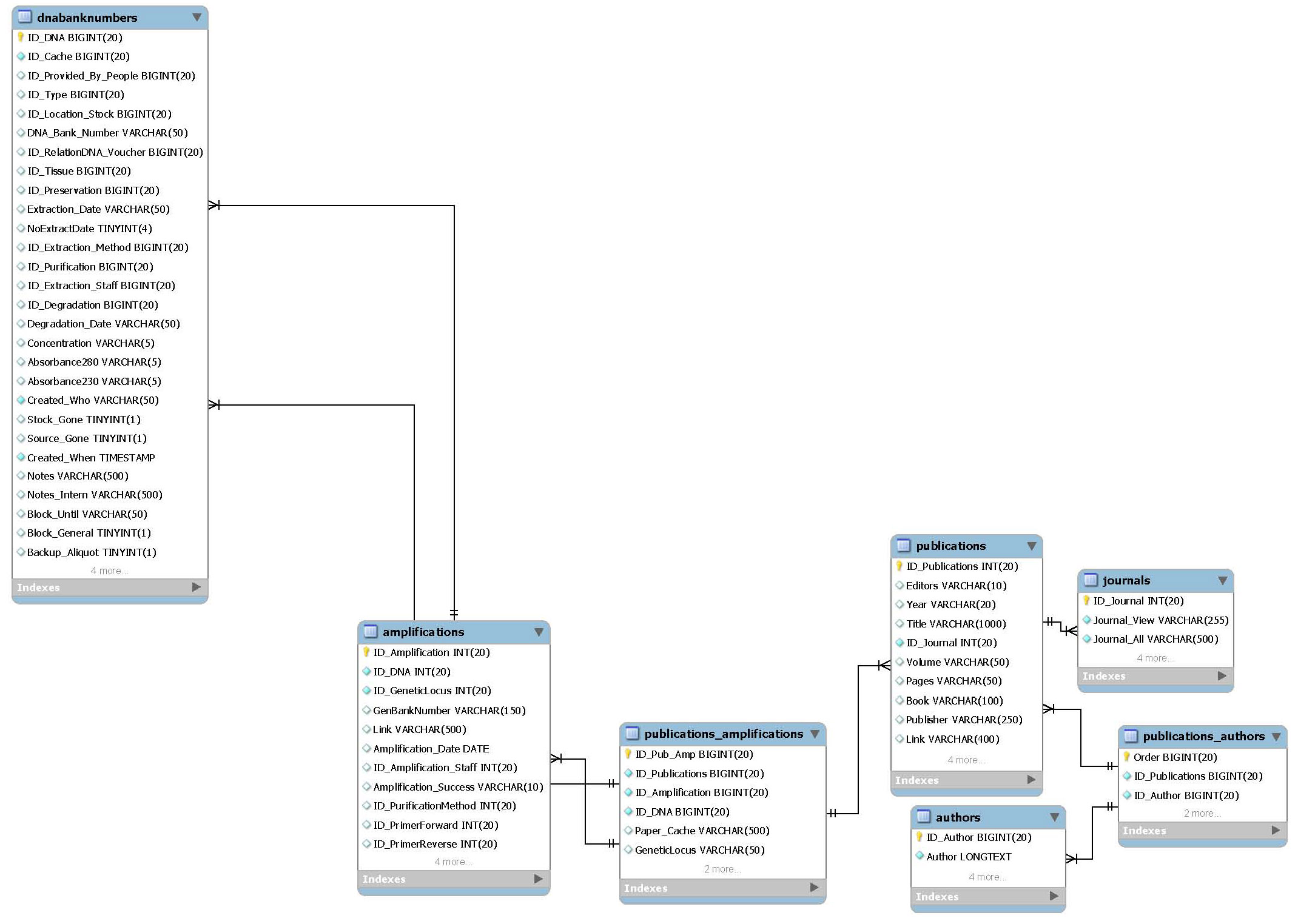
This package allows to associate molecular publications to DNA samples and sequence data. It should not be a complete substitute for a literature management software.
- Main table:
- publications
- Related packages:
- DNA extractions
- Sequence data
Sequence data
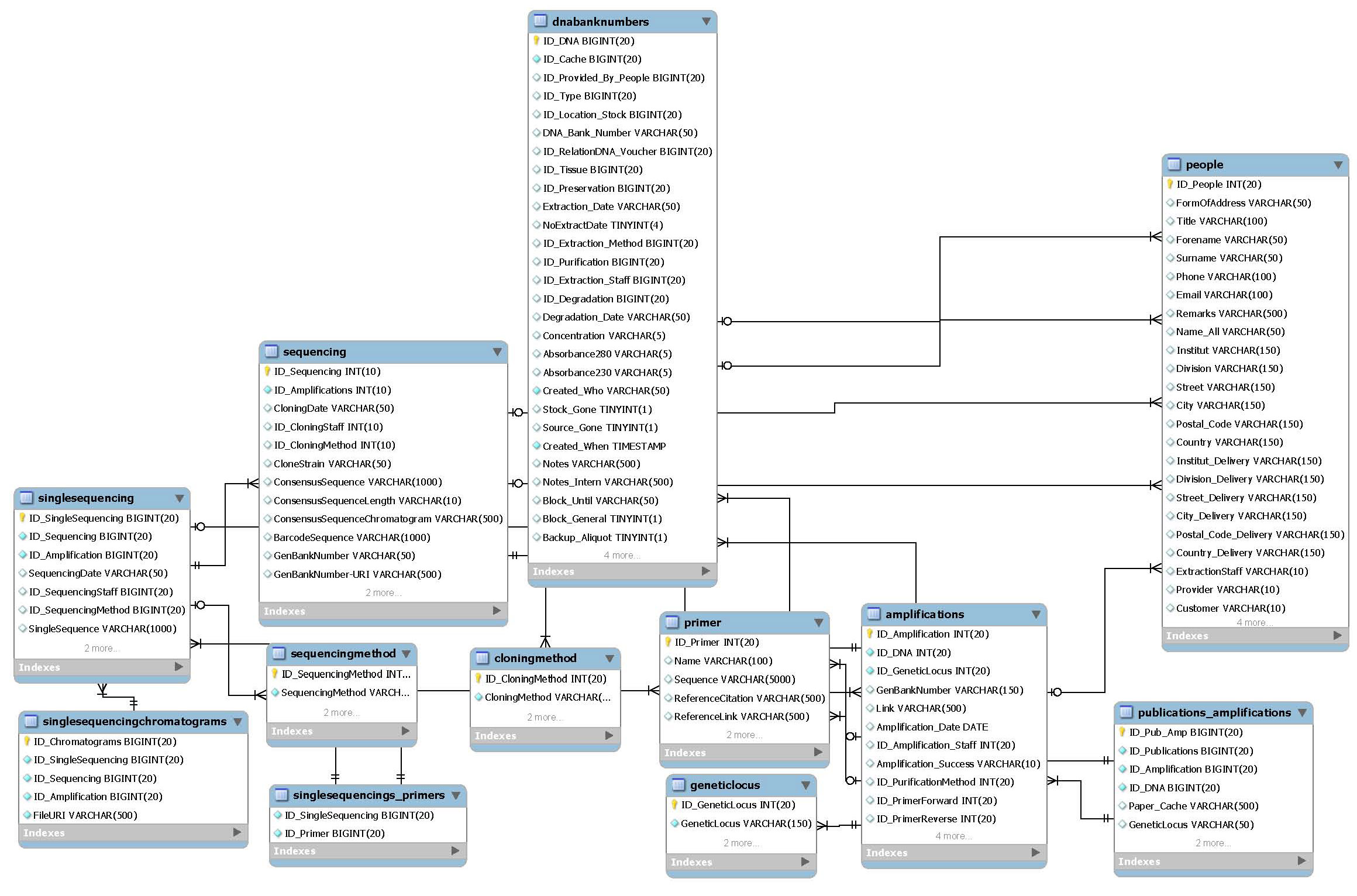
This package is for management of GenBank Accession Numbers, BOLD process IDs and Amplifications in general. More tables have been modelled for future management of raw sequence data, including primers, references, cloning etc. The frontend for this tables will be ready for use until end of 2011. For now only GenBank/BOLD accessions and Amplifications can be administrated.
- Main tables:
- amplifications
- sequencing
- List tables:
- geneticlocus
- primer
- cloningmethod
- sequencingmethod
- Related packages:
- DNA extractions
- Molecular publications
Customer requests
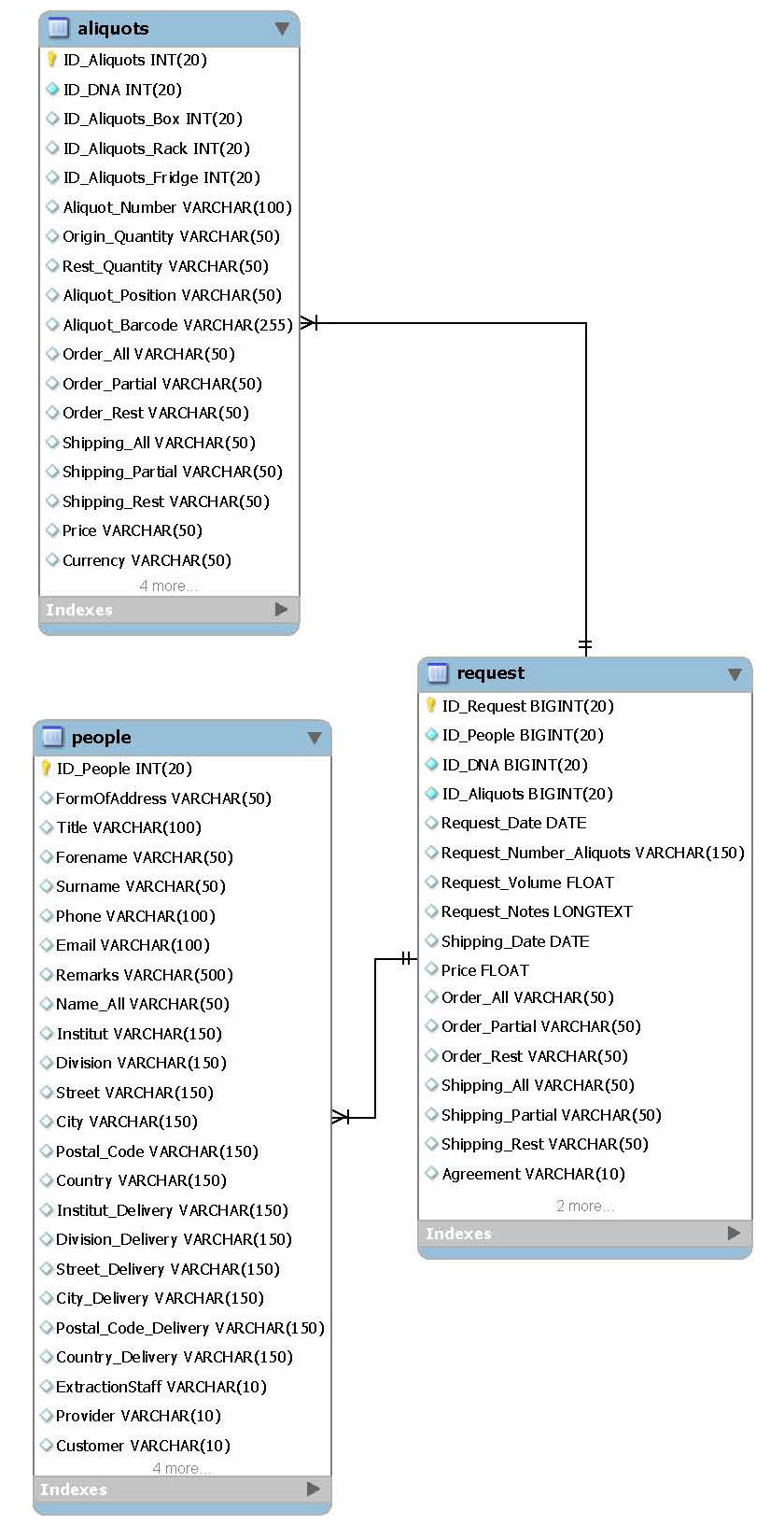
This package includes management of all customer requests respectively orders. Since way of administer payments depends on institution this package is for the lab site only.
- Main table:
- request
- Related packages:
- Location stock/aliquots
- DNA extractions
Specimen Tool
DNA Bank Network
Users
Log tables
Kurze Einführung, worum gehts, Allgemeiner Aufbau