Difference between revisions of "Installation"
(→Updating a regular BioCASe installtion) |
(→Installing BioCASe from a Docker image) |
||
| (59 intermediate revisions by the same user not shown) | |||
| Line 8: | Line 8: | ||
# A direct connection can be established from this machine to the database to be published – either because there’s no firewall in between, or the port used by the DBMS is open (e. g. 3306 for MySQL). | # A direct connection can be established from this machine to the database to be published – either because there’s no firewall in between, or the port used by the DBMS is open (e. g. 3306 for MySQL). | ||
| − | In addition to the regular installation | + | In addition to the regular installation, you can install BioCASe from a Docker image. Docker is an operating-system-level virtualization software that allows easy software deployment in so-called containers. If you don't know yet what Docker is, you should read the [https://docs.docker.com/get-started/ Get Started Guide]. '''If you're famliar with Docker, we'd suggest to use the Docker path for installing BioCASe''' - read more in section [[Installation#Installing_BioCASe_from_a_Docker_image | Installing from a Docker image]] below.''' |
| + | |||
| + | == Installing BioCASe from a Docker image == | ||
| + | |||
| + | Good choice! Installing BioCASe from a Docker image is much easer than installing the required components step-by-step. In addition, it's easier to update an existing installation. If you don't know yet what Docker is, you should read the [https://docs.docker.com/get-started/ Get Started Guide] first. | ||
| + | |||
| + | '''Important note:''' BioCASe requires Docker version 17 or later. If you already have Docker installed, run ''docker --version'' in a terminal and make sure the version number is 17+. | ||
| + | |||
| + | For installing BioCASe from the Docker image, please follow these steps: | ||
| + | |||
| + | # If you're not using Linux, install the ''make'' tool from https://gnuwin32.sourceforge.net/packages/make.htm. | ||
| + | # Install Docker Community Edition (Docker CE). For detailed instructions, see [https://docs.docker.com/install/ About Docker CE] and choose the tutorial for your platform. To make sure Docker is properly installed, run ''docker --version'' in a terminal and make sure the version number is 17+. | ||
| + | # Download the [http://ww2.biocase.org/svn/bps2/trunk/Makefile Makefile] that contains the commands for installing and updating BioCASe. Use "Save as..." to store it to your home folder on the server machine. | ||
| + | # Open a terminal window and change the work directory to the location you've just saved the Makefile to. Then type in ''make install'' and press enter. This should download the latest BioCASe image, create a BioCASe container and run it. | ||
| + | |||
| + | Per default, Docker will use port 80 for BioCASe. If this port is already used by another application on your server, you can instruct Docker to use another port. For using port 8181, for example, type in ''make install PORT=8181'' instead: | ||
| + | |||
| + | [[File:DockerInstall.png|650px]] | ||
| + | |||
| + | Voilà, that's it! To check your BioCASe installation, use your Browser to go to ''http://<dockerhost>'' (or ''http://<dockerhost>:8181'' if you used port 8181). Don't forget to [[Installation#Changing_the_Default_Password | change the default password]]. | ||
| + | |||
| + | You can also use the Makefile for stopping and starting the BioCASe container (for example, if you restart your server each night automatically) - just type ''make stop'' and ''make start''. For removing the container, use ''make remove'', for updating your BioCASe installation ''make update''. | ||
| + | |||
| + | '''Tell BioCASe your dockerhost''' Last thing to do is to tell BioCASe the name of your dockerhost: On the start page of BioCASe, choose <small>Config Tool</small>, then <small>System Administration</small>. Enter the password you just set and click <small>Submit</small>. Type in the URL you just used in your browser (''http://<dockerhost>'' or ''http://<dockerhost>:8181'' if you used port 8181) in the first line as <small>Webserver domain</small>. | ||
| + | |||
| + | '''Important note:''' The configuration and the log files of your BioCASe installation, as well as any XML and DwC archives created, will be stored in Docker hosted volumes. That means they are stored outside of the BioCASe container and will be preserved, even when the container is removed (actually, that's what happens if you update your installation). Docker hosted volumes are stored in the ''volumes'' folder of your docker installation, e.g. /var/lib/docker/volumes. If you want to backup the configuration of your BioCASe installation, just backup the folder /var/lib/docker/volumes/bps_config. To learn more about docker hosted volumes, please refer to the [https://docs.docker.com/storage/volumes/ Docker guide]. | ||
== Regular BioCASe Installation == | == Regular BioCASe Installation == | ||
| Line 15: | Line 40: | ||
BioCASe runs on both Windows and Linux machines. Two thoughts to consider for you: | BioCASe runs on both Windows and Linux machines. Two thoughts to consider for you: | ||
* If you want to use ODBC to connect to your database (for example to an Access database), you should make sure that an ODBC driver is available for the OS of your choice. For an Access database, for example, such a ODBC driver is available only for Windows - so you should go for Windows. | * If you want to use ODBC to connect to your database (for example to an Access database), you should make sure that an ODBC driver is available for the OS of your choice. For an Access database, for example, such a ODBC driver is available only for Windows - so you should go for Windows. | ||
| − | * If you don't wanna use ODBC, | + | * If you don't wanna use ODBC, because you plan to connect to a wide-spread DB system such as MySQL, Postgres or MS SQL Server, we recommend Linux (Debian). |
=== Installing Apache/IIS and Python === | === Installing Apache/IIS and Python === | ||
| Line 22: | Line 47: | ||
Even though the BPS runs with both Apache and Internet Information Server (IIS), we recommend using Apache, which is much easier to configure. However, if you’re already using IIS, you might want to stick with that. | Even though the BPS runs with both Apache and Internet Information Server (IIS), we recommend using Apache, which is much easier to configure. However, if you’re already using IIS, you might want to stick with that. | ||
| − | The BPS requires Python | + | The BPS requires Python 3.7+ to run, it doesn't work with Python 2, which ended its life cycle in 2020 (same is true for Python 3 versions < 3.7). On most Linux systems, Python 3 will be pre-installed. If not, you should be able to install Python using ''apt-get update'' and ''apt-get install python''. (By the way: You can install several different Python versions on the same machine without any conflicts.) |
If you’re installing on a Windows Vista (poor you) or Windows 7 machine, we recommend not to install Apache in the default program files folder, since there is some magic spell on that which can get you into trouble when changing Apache’s configuration files. Just choose another path (Apache Software Foundation?) to spare that hassle. | If you’re installing on a Windows Vista (poor you) or Windows 7 machine, we recommend not to install Apache in the default program files folder, since there is some magic spell on that which can get you into trouble when changing Apache’s configuration files. Just choose another path (Apache Software Foundation?) to spare that hassle. | ||
| Line 28: | Line 53: | ||
Once you’ve successfully installed Apache and Python, you can proceed to the next step. | Once you’ve successfully installed Apache and Python, you can proceed to the next step. | ||
| − | === Downloading the Provider Software === | + | === Downloading and unpacking the Provider Software === |
| − | You can | + | You can find the latest release of BioCASe on our [https://git.bgbm.org/biocase/bps/-/releases/permalink/latest Git page]. There are two archives available for download: |
| − | + | * The [https://git.bgbm.org/biocase/bps/-/archive/3.8.6/bps-3.8.6.zip default package (recommended)] contains the full installation and allows the generation of DarwinCore archives. For this feature, the [https://github.com/pentaho/pentaho-kettle Kettle libraries] are included in the download. | |
| − | + | * The [https://www.biocase.org/products/provider_software/files/bps-3.8.6-minimal.zip minimal package] doesn't contain the Kettle libraries, hence it's not possible to create DarwinCore archives with such an installation. Choose this download only if you have a good reason to do so. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | The | ||
| − | + | Download the file to your server and unpack it to the desired location of your file system. It contains the BioCASe Provider Software in a folder that contains the version number, e.g. ''bps-3.8.0''. We recommend to rename it to something that doesn't include the version number, so you don't need to touch your web server's configuration during future updates. | |
| − | |||
| − | |||
For the remainder of this guide, we will assume you’ve installed the BPS into a folder named ''biocase'', which should look like this by now: | For the remainder of this guide, we will assume you’ve installed the BPS into a folder named ''biocase'', which should look like this by now: | ||
| − | [[image:instBiocaseFolder.png| | + | [[image:instBiocaseFolder.png|650px]] |
=== Running the Setup Script === | === Running the Setup Script === | ||
| Line 52: | Line 69: | ||
In order to run the script, open a terminal window (or “command prompt” in Windows). Change the working directory to the folder you’ve downloaded the BPS to (''biocase'') and type in:<pre>python setup.py</pre> In case Python is not found, you’ll have to add the full path name to the Python executable. Make sure the Python version given in the fifth line of the output is the one you desire (otherwise add the path to the correct executable): | In order to run the script, open a terminal window (or “command prompt” in Windows). Change the working directory to the folder you’ve downloaded the BPS to (''biocase'') and type in:<pre>python setup.py</pre> In case Python is not found, you’ll have to add the full path name to the Python executable. Make sure the Python version given in the fifth line of the output is the one you desire (otherwise add the path to the correct executable): | ||
| − | [[image:instSetupScriptStart.png| | + | [[image:instSetupScriptStart.png|550px]] |
The questions asked by the setup script do have defaults. If you want to keep them, just press <small>Return</small>. | The questions asked by the setup script do have defaults. If you want to keep them, just press <small>Return</small>. | ||
| Line 62: | Line 79: | ||
After answering these two questions, the script will update your files. At the end of the output you can find the lines you'll need later to create the link between BioCASe and your web server, so do not close the window! | After answering these two questions, the script will update your files. At the end of the output you can find the lines you'll need later to create the link between BioCASe and your web server, so do not close the window! | ||
| − | [[image:instSetupScriptDone.png| | + | [[image:instSetupScriptDone.png|550px]] |
=== Linking BioCASe and Your Web Server === | === Linking BioCASe and Your Web Server === | ||
| Line 71: | Line 88: | ||
If you use Apache, adding the BioCASe folder to your web server environment is as simple as copying the lines printed at the end of the setup script to the end of the Apache configuration file ''httpd.conf''. This can typically be found in the ''conf'' folder of your Apache installation. If you’re adding BioCASe to an already productive web server, please ask the server admin before doing any changes! | If you use Apache, adding the BioCASe folder to your web server environment is as simple as copying the lines printed at the end of the setup script to the end of the Apache configuration file ''httpd.conf''. This can typically be found in the ''conf'' folder of your Apache installation. If you’re adding BioCASe to an already productive web server, please ask the server admin before doing any changes! | ||
| − | [[image:instHttpdConf.png| | + | [[image:instHttpdConf.png|550px]] |
Save the file and restart Apache for the changes to take effect. | Save the file and restart Apache for the changes to take effect. | ||
| Line 86: | Line 103: | ||
* http://haishibai.blogspot.de/2011/02/setting-up-python-on-iis-7_01.html | * http://haishibai.blogspot.de/2011/02/setting-up-python-on-iis-7_01.html | ||
* http://support.microsoft.com/kb/276494 | * http://support.microsoft.com/kb/276494 | ||
| − | |||
==== Testing the Link ==== | ==== Testing the Link ==== | ||
| Line 92: | Line 108: | ||
Once you’re done, you can test your BioCASe Installation by opening a browser and typing in ''http://localhost/biocase'' (replace ''localhost'' with the domain name of the machine you installed BioCASe on, if you’re not working locally). You should then see the start page of the BPS: | Once you’re done, you can test your BioCASe Installation by opening a browser and typing in ''http://localhost/biocase'' (replace ''localhost'' with the domain name of the machine you installed BioCASe on, if you’re not working locally). You should then see the start page of the BPS: | ||
| − | [[image:instBiocaseStartpage.png| | + | [[image:instBiocaseStartpage.png|550px]] |
| − | === Installing Additional Python Packages/Java === | + | === Installing Additional Python Packages/Java/Graphviz === |
Before you can start configuring your installation, you need to install additional packages, depending on the management system used for the database to be published and the functions of the BPS you intend to use. | Before you can start configuring your installation, you need to install additional packages, depending on the management system used for the database to be published and the functions of the BPS you intend to use. | ||
| − | On the BPS start page, click on <small>Utilities</small>, then <small>library test</small> in order to navigate to the library test page that shows the status of installed libraries. On a fresh BPS installation, | + | On the BPS start page, click on <small>Utilities</small>, then <small>library test</small> in order to navigate to the library test page that shows the status of installed libraries. On a fresh BPS installation, only the BioCASe libraries should be installed, since they're part of the archive file you’ve downloaded (or the Subversion repository you checked out from): |
| − | [[image:instLibrariesVanilla.png| | + | [[image:instLibrariesVanilla.png|550px]] |
| − | + | If you plan to use the Local Query Tool (a small portal for querying and browsing your BioCASe webservice), you need to install the lxml library. You can do this easily by running the Python Package Installer (PIP): ''pip install lxml''. | |
| − | For each DBMS you plan to use with BioCASe (i.e. you have a database you want to publish with BioCASe), you’ll have to install the corresponding package. So if your collection is stored in a MySQL database, | + | For each DBMS you plan to use with BioCASe (i.e. you have a database you want to publish with BioCASe), you’ll have to install the corresponding package. So if your collection is stored in a MySQL database, you'll need the MySQLdb library; the table shows you the command to run for installing it using pip: ''pip install mysqlclient''. Once pip has installed the package, reload the libraries test page. The according list entry should turn from an orange ''not installed'' into a green version number. Redo this process for all DBMS you want to connect to. |
| − | The Graphviz library listed under optional external binaries is used by the BPS to create a graph of your database’s data model in the table setup (you will learn in the [[DatasourceSetup#Setting_up_the_Database_Structure|Table Setup]] about this). If the data model is very simple (i.e., not more than 3 or 4 tables), you probably can spare the effort of installing Graphviz. Otherwise we recommend installing it: Simply click on the Download link | + | The Graphviz library listed under optional external binaries is used by the BPS to create a graph of your database’s data model in the table setup (you will learn in the [[DatasourceSetup#Setting_up_the_Database_Structure|Table Setup]] about this). If the data model is very simple (i.e., not more than 3 or 4 tables), you probably can spare the effort of installing Graphviz. Otherwise we recommend installing it: Simply click on the Download link and install the package using an installer file or a package manager on Linux. |
| − | If you want to create DarwinCore Archives for your web services, you also need to install a Java Runtime Environment (JRE). | + | If you want to create DarwinCore Archives for your web services, you also need to install a Java Runtime Environment (JRE). BioCASe requires Java 1.8 or later for creating DarwinCore archives, so please check the version listed on the Testlibs page if it's already installed on the server. |
=== Changing the Default Password === | === Changing the Default Password === | ||
| Line 122: | Line 138: | ||
In the first two lines, you’ll see the values you entered during the setup script (in this case, the defaults ''localhost'' and ''biocase''). In the fourth line, enter a new password. Please be careful when typing (if you forget the password some day, you can reset it by directly editing the configuration file). Press <small>update config</small> to save the new password to the configuration file. | In the first two lines, you’ll see the values you entered during the setup script (in this case, the defaults ''localhost'' and ''biocase''). In the fourth line, enter a new password. Please be careful when typing (if you forget the password some day, you can reset it by directly editing the configuration file). Press <small>update config</small> to save the new password to the configuration file. | ||
| − | In case you’ve installed Graphviz, you need to tell the BPS the location of the binaries, since it is not a Python package that will be registered automatically. In order to do so, locate the Graphviz installation on your local disk and enter the path to the binary executable into the text box labelled <small>Graphviz | + | In case you’ve installed Graphviz, you need to tell the BPS the location of the binaries, since it is not a Python package that will be registered automatically. In order to do so, locate the Graphviz installation on your local disk and enter the path to the binary executable into the text box labelled <small>Path to Graphviz executable</small>. On Linux machines it might look similar to the default (''/usr/local/bin/dot''), on Windows machines it will be probably something like ''C:/Program Files/Graphviz 2.44.1/bin/''. Press <small>update config</small> to save the path. |
=== Checking Your Installation === | === Checking Your Installation === | ||
| Line 129: | Line 145: | ||
# Typing in ''http://localhost/biocase'' (or the appropriate values of your installation) takes you to the entry page of the provider software (as shown in the section [[#Linking_BioCASe_and_Your_Web_Server|Linking BioCASe and Your Web Server]]). | # Typing in ''http://localhost/biocase'' (or the appropriate values of your installation) takes you to the entry page of the provider software (as shown in the section [[#Linking_BioCASe_and_Your_Web_Server|Linking BioCASe and Your Web Server]]). | ||
| − | # The libraries test page (<small>Utilities > library test</small>) shows you’ve installed the packages for | + | # The libraries test page (<small>Utilities > library test</small>) shows you’ve installed the packages for your DBMS, lxml (if you want to use the Local Query Tool), Java (if you want to create DarwinCore Archives) and the Graphviz dot (if you decided so). |
# <small>Status of writeable directories</small> shows a green <small>writeable</small> for all four directories. If not, the path to problematic file will be given – please grant write access to Python. | # <small>Status of writeable directories</small> shows a green <small>writeable</small> for all four directories. If not, the path to problematic file will be given – please grant write access to Python. | ||
| − | + | If you want to connect BioCASe to a variety of different database management systems, create DarwinCore archives and use the QueryTool, the libraries test pages should look like this: | |
| − | [[image:instLibrariesDone.png| | + | [[image:instLibrariesDone.png|650px]] |
| − | == | + | == Updating an existing BioCASe Installation == |
| − | == Updating | + | === Updating a BioCASe Docker installation === |
| + | This is very simple with the [http://ww2.biocase.org/svn/bps2/trunk/Makefile Makefile] you downloaded for installing BioCASe. Just go to the folder where you stored the file and run ''make update''. This will stop and remove the BioCASe container, download the latest BioCASe image and create/start a new BioCASe container. Since the configuration files are not stored within the container, all your data sources will be preserved. | ||
=== Updating a regular BioCASe installation === | === Updating a regular BioCASe installation === | ||
| Line 160: | Line 177: | ||
# Last, copy the configuration of the BioCASe web services: Open the folder ''config/datasources'' of your old installation and copy all subfolders to ''config/datasources'' of your new installation. | # Last, copy the configuration of the BioCASe web services: Open the folder ''config/datasources'' of your old installation and copy all subfolders to ''config/datasources'' of your new installation. | ||
# Done! [[Installation#Checking_Your_Installation|Try out your new installation]] to make sure it is working. | # Done! [[Installation#Checking_Your_Installation|Try out your new installation]] to make sure it is working. | ||
| − | |||
| − | |||
Latest revision as of 12:53, 5 August 2025
This part of the wiki explains how to install the BioCASe Provider Software. Depending on your skills, it should take between 15 and 60 minutes. If you’re a beginner, please read the Beginner’s Guide first.
Contents
- 1 Requirements and Considerations
- 2 Installing BioCASe from a Docker image
- 3 Regular BioCASe Installation
- 4 Updating an existing BioCASe Installation
Requirements and Considerations
Before installing, please make sure that the system you’re intending to install the BPS on meets the following requirements (If you find out one of these items is not met, please ask your system administrator for help, read the FAQ or get in contact with the BioCASe Team.):
- You have administrative privileges on this machine, i.e. the user rights to install new software packages.
- It is accessible from outside your institution. This means it either needs to be outside any institutional firewall or, more likely, the HTTP port is open for an existing firewall.
- A direct connection can be established from this machine to the database to be published – either because there’s no firewall in between, or the port used by the DBMS is open (e. g. 3306 for MySQL).
In addition to the regular installation, you can install BioCASe from a Docker image. Docker is an operating-system-level virtualization software that allows easy software deployment in so-called containers. If you don't know yet what Docker is, you should read the Get Started Guide. If you're famliar with Docker, we'd suggest to use the Docker path for installing BioCASe - read more in section Installing from a Docker image below.
Installing BioCASe from a Docker image
Good choice! Installing BioCASe from a Docker image is much easer than installing the required components step-by-step. In addition, it's easier to update an existing installation. If you don't know yet what Docker is, you should read the Get Started Guide first.
Important note: BioCASe requires Docker version 17 or later. If you already have Docker installed, run docker --version in a terminal and make sure the version number is 17+.
For installing BioCASe from the Docker image, please follow these steps:
- If you're not using Linux, install the make tool from https://gnuwin32.sourceforge.net/packages/make.htm.
- Install Docker Community Edition (Docker CE). For detailed instructions, see About Docker CE and choose the tutorial for your platform. To make sure Docker is properly installed, run docker --version in a terminal and make sure the version number is 17+.
- Download the Makefile that contains the commands for installing and updating BioCASe. Use "Save as..." to store it to your home folder on the server machine.
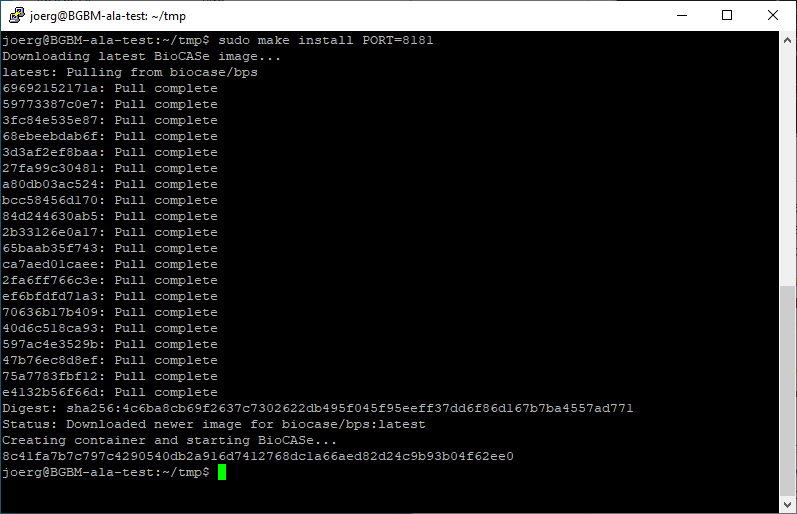
- Open a terminal window and change the work directory to the location you've just saved the Makefile to. Then type in make install and press enter. This should download the latest BioCASe image, create a BioCASe container and run it.
Per default, Docker will use port 80 for BioCASe. If this port is already used by another application on your server, you can instruct Docker to use another port. For using port 8181, for example, type in make install PORT=8181 instead:
Voilà, that's it! To check your BioCASe installation, use your Browser to go to http://<dockerhost> (or http://<dockerhost>:8181 if you used port 8181). Don't forget to change the default password.
You can also use the Makefile for stopping and starting the BioCASe container (for example, if you restart your server each night automatically) - just type make stop and make start. For removing the container, use make remove, for updating your BioCASe installation make update.
Tell BioCASe your dockerhost Last thing to do is to tell BioCASe the name of your dockerhost: On the start page of BioCASe, choose Config Tool, then System Administration. Enter the password you just set and click Submit. Type in the URL you just used in your browser (http://<dockerhost> or http://<dockerhost>:8181 if you used port 8181) in the first line as Webserver domain.
Important note: The configuration and the log files of your BioCASe installation, as well as any XML and DwC archives created, will be stored in Docker hosted volumes. That means they are stored outside of the BioCASe container and will be preserved, even when the container is removed (actually, that's what happens if you update your installation). Docker hosted volumes are stored in the volumes folder of your docker installation, e.g. /var/lib/docker/volumes. If you want to backup the configuration of your BioCASe installation, just backup the folder /var/lib/docker/volumes/bps_config. To learn more about docker hosted volumes, please refer to the Docker guide.
Regular BioCASe Installation
This is the path to choose if you haven't heard of Docker yet. If you have experience with this virtualisation software, we'd suggest to install BioCASe from the Docker image (instructions in section Installing from a Docker image).
BioCASe runs on both Windows and Linux machines. Two thoughts to consider for you:
- If you want to use ODBC to connect to your database (for example to an Access database), you should make sure that an ODBC driver is available for the OS of your choice. For an Access database, for example, such a ODBC driver is available only for Windows - so you should go for Windows.
- If you don't wanna use ODBC, because you plan to connect to a wide-spread DB system such as MySQL, Postgres or MS SQL Server, we recommend Linux (Debian).
Installing Apache/IIS and Python
If you’re using a Linux system, you probably don’t need to install either, since Apache and Python are usually preinstalled. Same holds true for MacOS, but you should have a look at one of the articles on the web to find out about the file locations.
Even though the BPS runs with both Apache and Internet Information Server (IIS), we recommend using Apache, which is much easier to configure. However, if you’re already using IIS, you might want to stick with that.
The BPS requires Python 3.7+ to run, it doesn't work with Python 2, which ended its life cycle in 2020 (same is true for Python 3 versions < 3.7). On most Linux systems, Python 3 will be pre-installed. If not, you should be able to install Python using apt-get update and apt-get install python. (By the way: You can install several different Python versions on the same machine without any conflicts.)
If you’re installing on a Windows Vista (poor you) or Windows 7 machine, we recommend not to install Apache in the default program files folder, since there is some magic spell on that which can get you into trouble when changing Apache’s configuration files. Just choose another path (Apache Software Foundation?) to spare that hassle.
Once you’ve successfully installed Apache and Python, you can proceed to the next step.
Downloading and unpacking the Provider Software
You can find the latest release of BioCASe on our Git page. There are two archives available for download:
- The default package (recommended) contains the full installation and allows the generation of DarwinCore archives. For this feature, the Kettle libraries are included in the download.
- The minimal package doesn't contain the Kettle libraries, hence it's not possible to create DarwinCore archives with such an installation. Choose this download only if you have a good reason to do so.
Download the file to your server and unpack it to the desired location of your file system. It contains the BioCASe Provider Software in a folder that contains the version number, e.g. bps-3.8.0. We recommend to rename it to something that doesn't include the version number, so you don't need to touch your web server's configuration during future updates.
For the remainder of this guide, we will assume you’ve installed the BPS into a folder named biocase, which should look like this by now:
Running the Setup Script
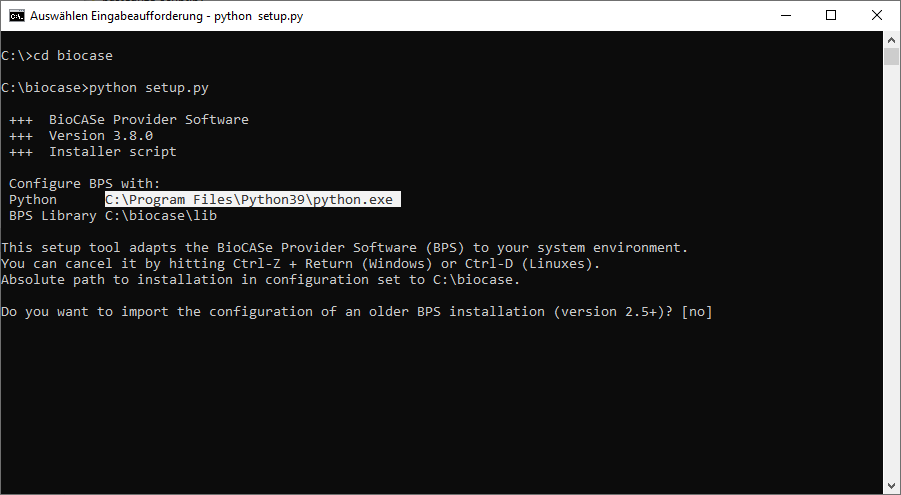
The setup script will initiate the configuration files with values you provide and point the script files to your Python installation (you can re-run the setup script in case that location changes). If you want to modify any of settings you made during the setup, you can change the values in the configuration tool later.
In order to run the script, open a terminal window (or “command prompt” in Windows). Change the working directory to the folder you’ve downloaded the BPS to (biocase) and type in:python setup.pyIn case Python is not found, you’ll have to add the full path name to the Python executable. Make sure the Python version given in the fifth line of the output is the one you desire (otherwise add the path to the correct executable):
The questions asked by the setup script do have defaults. If you want to keep them, just press Return.
Webserver domain: This is the domain name of the web server you’re installing BioCASe on, e.g. http://www.myinstitution.org. You can keep the default localhost as long as you’re configuring and testing the installation locally; for production you’ll need to change that into the correct domain name (or IP address) later.
Base URL: This will become the second part of the URL your installation will use. If you keep the default biocase, your installation will be accessible later at www.myinstitution.org/biocase, for example.
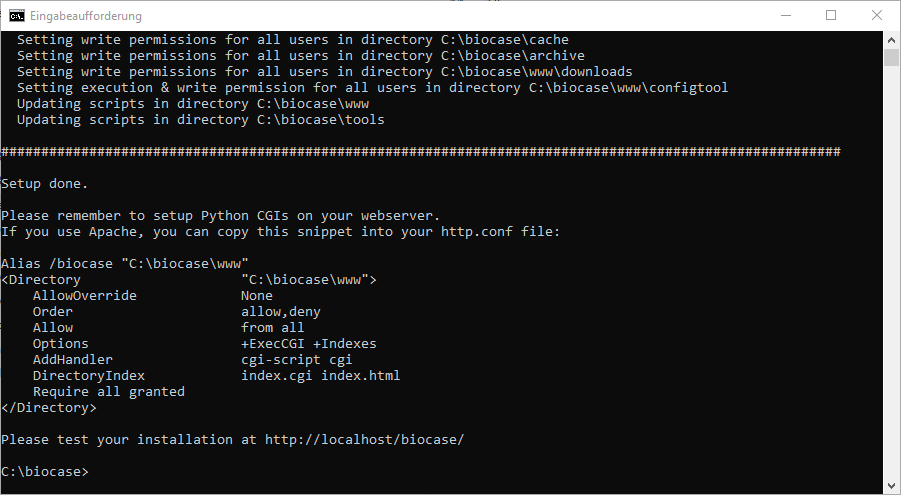
After answering these two questions, the script will update your files. At the end of the output you can find the lines you'll need later to create the link between BioCASe and your web server, so do not close the window!
Linking BioCASe and Your Web Server
The public part of the provider software is stored in the www subfolder of your BioCASe installation and must be configured in your web server as a public folder. You should always make this folder public using a virtual directory: This means that you should never install BioCASe under the default root folder of your web server. Doing so can compromise your server security and will allow people to see your configuration files (which include passwords). So since we installed BioCASe in c:\biocase, the folder that must be published with a virtual directory is c:\biocase\www and will be accessible later through http://www.myinstitution.org/biocase.
Apache
If you use Apache, adding the BioCASe folder to your web server environment is as simple as copying the lines printed at the end of the setup script to the end of the Apache configuration file httpd.conf. This can typically be found in the conf folder of your Apache installation. If you’re adding BioCASe to an already productive web server, please ask the server admin before doing any changes!
Save the file and restart Apache for the changes to take effect.
Internet Information Server
If you use Internet Information Server, you need to create a virtual directory for the folder c:\biocase\www using the IIS Server Manager. Grant all rights for this directory (Read, Write, Script Execution, Directory listing). You should also add index.html to the list of default documents (Virtual Directory > Properties > Documents > Default).
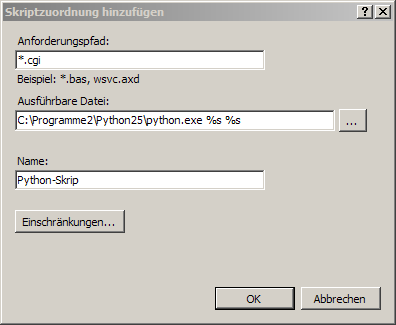
In order to associate Python scripts with Python in IIS 5/6, you need to use the Configure button on the virtual directory tab and associate the file extensions .cgi and .py with Python (see screenshot below). In IIS 7, you can open this dialog by choosing Add Script Map. Note the placeholders %s %s after the Python executable!
If you run into problems configuring IIS, you should have a look at the tutorials available on the web, for example
- http://haishibai.blogspot.de/2011/02/setting-up-python-on-iis-7_01.html
- http://support.microsoft.com/kb/276494
Testing the Link
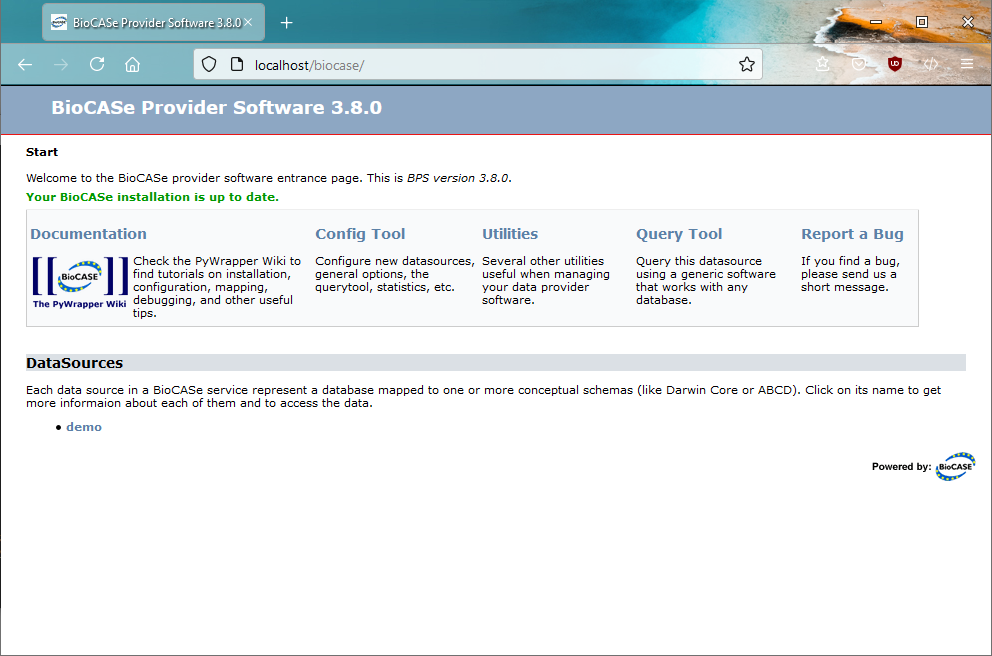
Once you’re done, you can test your BioCASe Installation by opening a browser and typing in http://localhost/biocase (replace localhost with the domain name of the machine you installed BioCASe on, if you’re not working locally). You should then see the start page of the BPS:
Installing Additional Python Packages/Java/Graphviz
Before you can start configuring your installation, you need to install additional packages, depending on the management system used for the database to be published and the functions of the BPS you intend to use.
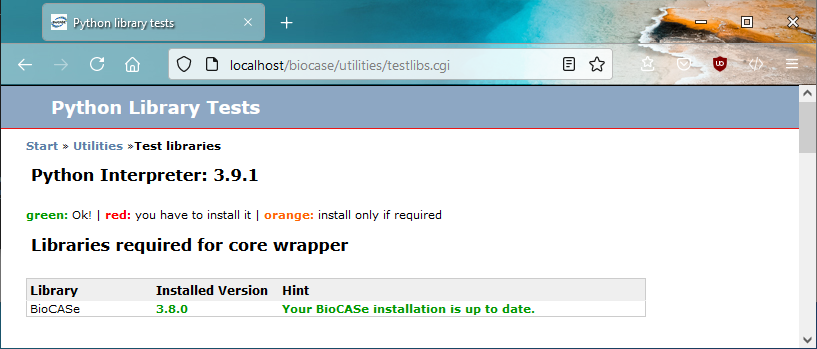
On the BPS start page, click on Utilities, then library test in order to navigate to the library test page that shows the status of installed libraries. On a fresh BPS installation, only the BioCASe libraries should be installed, since they're part of the archive file you’ve downloaded (or the Subversion repository you checked out from):
If you plan to use the Local Query Tool (a small portal for querying and browsing your BioCASe webservice), you need to install the lxml library. You can do this easily by running the Python Package Installer (PIP): pip install lxml.
For each DBMS you plan to use with BioCASe (i.e. you have a database you want to publish with BioCASe), you’ll have to install the corresponding package. So if your collection is stored in a MySQL database, you'll need the MySQLdb library; the table shows you the command to run for installing it using pip: pip install mysqlclient. Once pip has installed the package, reload the libraries test page. The according list entry should turn from an orange not installed into a green version number. Redo this process for all DBMS you want to connect to.
The Graphviz library listed under optional external binaries is used by the BPS to create a graph of your database’s data model in the table setup (you will learn in the Table Setup about this). If the data model is very simple (i.e., not more than 3 or 4 tables), you probably can spare the effort of installing Graphviz. Otherwise we recommend installing it: Simply click on the Download link and install the package using an installer file or a package manager on Linux.
If you want to create DarwinCore Archives for your web services, you also need to install a Java Runtime Environment (JRE). BioCASe requires Java 1.8 or later for creating DarwinCore archives, so please check the version listed on the Testlibs page if it's already installed on the server.
Changing the Default Password
The configuration tool is a browser interface that allows you to configure almost every aspect of the BPS. As a last step of the setup, you should change the password.
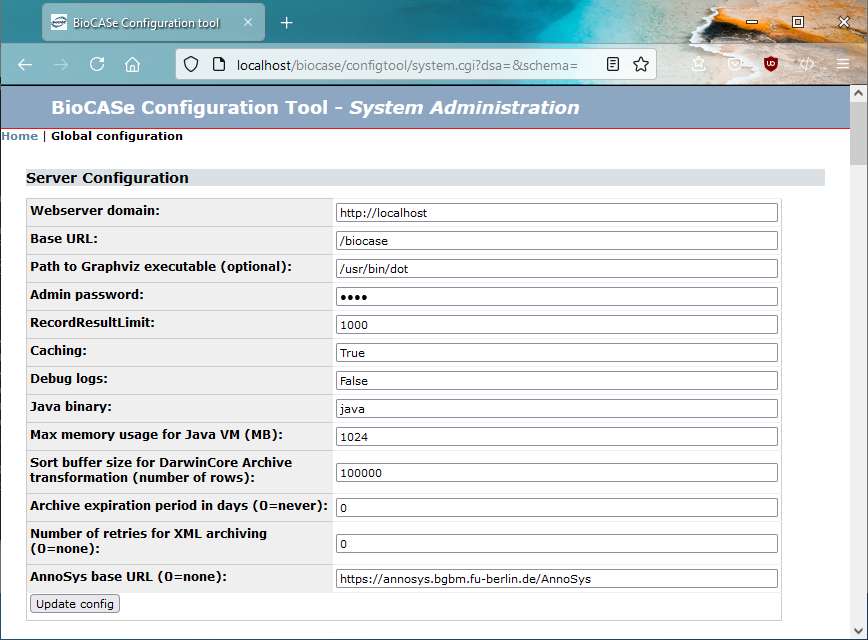
Go back to the Start page of the BPS (by clicking Start at the top of the page, or by retyping http://localhost/biocase if the server is running on your computer), choose Config Tool, then System Administration. You will then be prompted for a password. Type in the default ACDC, then click Submit.
The top of the configuration page you’ll be directed to should look similar to this:
In the first two lines, you’ll see the values you entered during the setup script (in this case, the defaults localhost and biocase). In the fourth line, enter a new password. Please be careful when typing (if you forget the password some day, you can reset it by directly editing the configuration file). Press update config to save the new password to the configuration file.
In case you’ve installed Graphviz, you need to tell the BPS the location of the binaries, since it is not a Python package that will be registered automatically. In order to do so, locate the Graphviz installation on your local disk and enter the path to the binary executable into the text box labelled Path to Graphviz executable. On Linux machines it might look similar to the default (/usr/local/bin/dot), on Windows machines it will be probably something like C:/Program Files/Graphviz 2.44.1/bin/. Press update config to save the path.
Checking Your Installation
Before you proceed to the configuration, you should make sure everything is installed correctly:
- Typing in http://localhost/biocase (or the appropriate values of your installation) takes you to the entry page of the provider software (as shown in the section Linking BioCASe and Your Web Server).
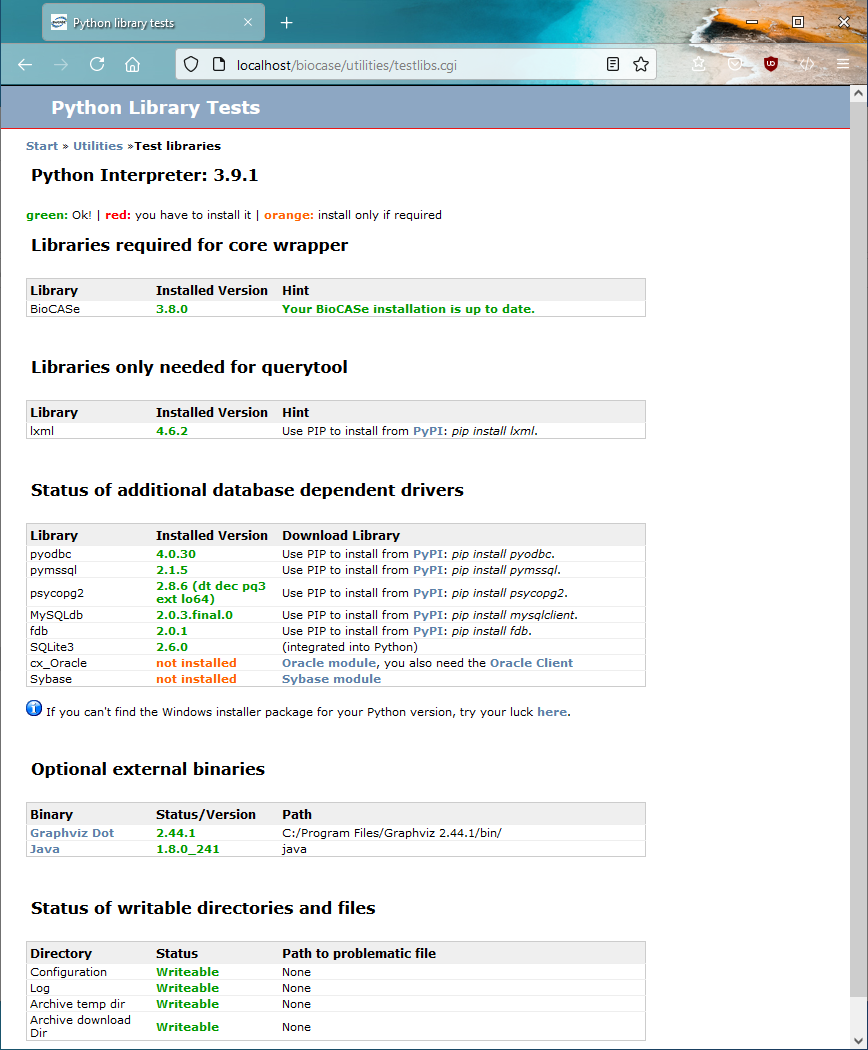
- The libraries test page (Utilities > library test) shows you’ve installed the packages for your DBMS, lxml (if you want to use the Local Query Tool), Java (if you want to create DarwinCore Archives) and the Graphviz dot (if you decided so).
- Status of writeable directories shows a green writeable for all four directories. If not, the path to problematic file will be given – please grant write access to Python.
If you want to connect BioCASe to a variety of different database management systems, create DarwinCore archives and use the QueryTool, the libraries test pages should look like this:
Updating an existing BioCASe Installation
Updating a BioCASe Docker installation
This is very simple with the Makefile you downloaded for installing BioCASe. Just go to the folder where you stored the file and run make update. This will stop and remove the BioCASe container, download the latest BioCASe image and create/start a new BioCASe container. Since the configuration files are not stored within the container, all your data sources will be preserved.
Updating a regular BioCASe installation
Important Note: If you have a running BioCASe installation, you shouldn’t touch it until you’ve successfully installed a new version. So keep the old installation instead of replacing it!
Updating from version 2.5 or later
- Rename the folder of the old installation to something meaningful, for example biocase_old, and memorise its old name. By doing this, you can always revert to this working version.
- Download the new version as described above into a directory with the name you just memorised.
- Change directory to the new installation and run setup script. When asked whether to import the configuration from an old installation, answer yes. The setup script will now import the configuration of the old installation, including the data sources.
- Done! Try out your new installation to make sure it is working.
Updating from older versions < 2.5
- Rename the folder of the old installation to something meaningful, for example biocase_old, and memorise its old name. By doing this, you can always revert to this working version; moreover, you will need the configuration files later.
- Download the new version as described above into a directory with the name you just memorised.
- Run the setup script. When asked whether to import the configuration from an old installation, just press Return (defaults to no). Make sure to provide the the same base URL that was used by the old installation (for example /biocase).
- Change the default password of the new installation to the one you used in the old installation.
- Open the file config/config.ini of your old installation in a text editor. Open the config tool of the new installation and navigate to the System Administration page. Compare the settings in the section Server configuration with the values you can find in the old configuration file and change them, if necessary (leave values unchanged if you don't find them in the old file). Press Update config.
- Last, copy the configuration of the BioCASe web services: Open the folder config/datasources of your old installation and copy all subfolders to config/datasources of your new installation.
- Done! Try out your new installation to make sure it is working.