Difference between revisions of "Archiving"
m (→DarwinCore Archives) |
(→Discovery of Archives) |
||
| (82 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | '''This page describes the archiving features in version 3. | + | '''This page describes the archiving features in version 3.4 or later.<br>If you're using version 3.0 or 3.1, please read the [[Archiving_v3.0/3.1|version 3.0/3.1 page]], for versions 3.2 and 3.3, go [http://wiki.bgbm.org/bps/index.php?title=Archiving&oldid=713 here].''' |
| − | With the archiving feature, you can create archive files that store all data published by your BioCASe web service. That might be handy if your database stores a huge number of records (several hundred | + | With the archiving feature, you can create archive files that store all data published by your BioCASe web service. That might be handy if your database stores a huge number of records (several hundred thousand or even millions) and the traditional harvesting approach of paging through the records published by the web service takes unacceptably long. Once stored in archives, these files can be downloaded from your web server or sent to the harvester via email and ingested much faster. The OpenUp! natural history aggregator, for example, can use this method for fast harvesting of huge datasets. |
| − | There are two types of archives: <strong>XML Archives</strong> simply store the records as compressed XML documents, each holding a customizable number of records. If your web service publishes 500,000 records by using the ABCD data schema, for example, an ABCD Archive created for that service could hold 500 ABCD documents, each storing 1,000 records. For each schema supported by a BioCASe web service, | + | There are two types of archives: <strong>XML Archives</strong> simply store the records as compressed XML documents, each holding a customizable number of records. If your web service publishes 500,000 records by using the ABCD data schema, for example, an ABCD Archive created for that service could hold 500 ABCD documents, each storing 1,000 records. For each schema supported by a BioCASe web service, separate XML archives can be created. You can download a sample ABCD Archive here: [[File:AlgenEngelsSmall_ABCD_2.06.zip]]. |
| − | + | The BioCASe Provider Software can create XML archives for each schema supported by a web service. For the ABCD 2.06 and HISPID 5 schemas, the XML Archiving is dataset-aware, that means each dataset published by the web service will end up in a separate archive file. All other schemas (including ABCD 1.2) will result in one single file for a given web service. | |
| − | + | <strong>DarwinCore Archives</strong> (DwCAs), in contrast, consist of one or several text files storing the core records and related information, zipped up together with a descriptor file and a metadata document. A detailed specification can be found on the [http://rs.tdwg.org/dwc/terms/guides/text/index.htm TDWG site], a sample archive here: [[File:Desmidiaceae_Engels.zip]]. | |
| + | |||
| + | Information published by web services using the ABCD 2.06 or HISPID 5 schema (dubbed as "ABCD/HISPID dumps") can be stored as DarwinCore Archives. Unsurprisingly, DwCAs make use of the [http://rs.tdwg.org/dwc/terms/index.htm DarwinCore data standard], which is flat and less complex than ABCD 2.06 or HISPID 5. Consequently, a DarwinCore archive will store less information than the corresponding XML archive for a given dataset. | ||
| + | |||
| + | ==Managing Archives== | ||
| + | The section <small>Existing Archives</small> allows you download, remove and convert archives or view the log of the archiving process: | ||
| + | |||
| + | [[File:xmlArchivesNew.png|border|775px]] | ||
| + | |||
| + | The archives are grouped by schema, with DarwinCore archives making up an additional group. You can use the minus or plus symbol next to the group header to expand or collapse all files of that group; for web services publishing dozens of datasets, this might be handy to reduce scrolling on the page. The <small>Log </small> link in the group header will open the log file for the respective archiving process in a separate tab. | ||
| + | |||
| + | For each archive file, the number of records stored in this file, the date of creation and the file size are listed. XML archives may have a number printed in the <small>Dropped</small> column, meaning some records have been dropped due to missing mandatory data elements. This doesn't mean the archive is invalid, but you should review the mapping or your source data to find the reasons for these lost records. Clicking the archive name will download the archive file, so it can be saved to disk or opened with an archive viewing tool. Pressing the trash symbol at the very right will delete the archive; if you use the trash symbol in the group header, all archives of this schema/type will be removed. | ||
| + | |||
| + | For ABCD 2.06 and HISPID 5 XML archives, you will see an additional <small>DwC</small> link next to the trash symbol. This allows you to selectively convert a single dataset from XML to DarwinCore format. In contrast to using the <small>Create DwC Archive</small> button described below, this will spare the XML archiving of the whole web service and just create a DarwinCore archive for the selected dataset, which will be significantly faster. If you use the link in the group header, all archives of the schema will be converted (again without triggering XML archiving first). | ||
==XML Archives== | ==XML Archives== | ||
| − | + | If you’ve finished mapping a schema (see [[ABCD2Mapping]] to learn how to map ABCD, for example) and tested the resulting web service, you can create XML Archives for that web service. To go to the archiving page, simply use the <small>Archive</small> link on the overview page of the datasource setup. If you haven’t mapped a schema, you won’t be able to create any archives. Before archiving, you should always make sure the web service returns correct results by using the QueryForms. | |
| + | |||
| + | The section <small>Create/Update Archives</small> allows you to create new XML archive(s) for each schema mapped. Simply select the schema and press <small>Create XML Archive</small>. As mentioned above, archiving ABCD 2.06 or HISPID 5 is dataset-aware, resulting in one XML archive per dataset published. For all other schemas, a single archive file will be created. | ||
| + | |||
| + | The paging size specifies how many records will be bundled into one XML document inside the archive; the default 1,000 should be suitable for most cases. Be aware that a BioCASe web service can restrict the number of records per request - if that limit is below the paging size set for archiving, the paging size will be adjusted automatically after the first packet of records was received from the web service. | ||
| + | |||
| + | During the archiving process, the <small>Log</small> will display messages indicating the progress: | ||
| + | |||
| + | [[File:xmlArchivesProgressNew.png|border]] | ||
| + | |||
| + | You can always cancel the archiving process by pressing the <small>Cancel</small> link in the Status/Messages box. However, it will take some time for the cancelling to show effect, namely until the current paging step has been completed. Depending upon the paging size set and the speed of your server, this can be up to one minute. | ||
| + | |||
| + | As the archiving progresses, the first lines of the log will drop out of view. Pressing <small>Show full log</small> will open the full log in a separate browser tab. You can navigate away from the archiving page during the process (or simply close it) and return later. Once finished, the dump will show up in the <small>Existing Archives</small> list, or, if it’s still being processed, as <small>Processing</small> in the Status/Messages box. If the process failed for some reason (or you decided to cancel it), a new box <small>Logs of aborted/failed runs</small> will be displayed with a link for displaying the log file. This will allow you to find out what caused the abortion. | ||
| + | |||
| + | For a given web service, you can only create XML archives for one schema at a time. Even though you can start archiving for several web services of your BioCASe installation in parallel, it is not a good idea. The archiving process will put heavy load on your servers – both the database server for data retrieval and the BioCASe server for creating the XML documents. So it is advisable to create just one archive at a time in order to allow both servers to respond to other requests. | ||
| + | |||
| + | ==DarwinCore Archives== | ||
| + | For web services that use the ABCD 2.06 or HISPID 5 schema, all information published by the web service can be stored in DarwinCore Archives. The DarwinCore data standard is less complex than ABCD, so in most cases the DarwinCore Archive will not store all information published by the web service. DarwinCore archives store only one dataset; so as with ABCD 2.06 and HISPID 5 XML archives, DwC-archiving a web service might result in several DarwinCore archives. | ||
| + | |||
| + | DarwinCore archives are not created directly, but from the respective ABCD/HISPID archives. So DwC-archiving a web service will first create ACBD archives, which will be converted into the respective DwC archives subsequently. | ||
| − | + | ===Preparation=== | |
| + | The conversion of ABCD into DwC is done through the Java-based Kettle library. Therefore, you need to have a Java Runtime Environment (version 1.5 or later) installed. You can check this on the library test page under ''Optional external binaries'': | ||
| − | + | [[File:dwcaJavaTest.png|border|500px]] | |
| − | |||
| − | + | If you don't have one, you can get it [http://www.java.com/de/download/ here]. Important: If you're running BioCASe on a 64bit machine, make sure to get a 64bit Java, since this will boost performance considerably. | |
| − | + | Once Java is installed, go to the System Administration (<small>Start > Config Tool > System Administration</small>) to check and adapt the conversion settings to your environment: | |
| − | + | [[File:dwcaSystemSettings.png|600px]] | |
| − | + | '''Java binary:''' If you're sure Java is installed, but the test lib page shows ''not installed'', please provide the full path to the Java binary (for example ''/usr/local/jdk1.6.0/bin/java''). If you have several Java versions installed (for example 32bit and 64bit versions), make sure to point to the correct (preferably 64bit) version. | |
| − | For | + | '''Max memory usage for Java VM (MB):''' This is the maximum amount of heap memory Java will be allowed to use (in MB). The default of 1024 will be sufficient for small datasets (up to 100,000 records). For medium-sized datasets (up to 1m records), you should set this value 2048. For large datasets with millions of records, a value of 4096 is recommended. If your server has enough free memory, you should be generous with this limit - larger values speed up the transformation process on most machines. However, you should make sure to leave enough memory for other required applications on your server. |
| − | + | '''Sort buffer size for transformation (number of rows):''' This is the number of rows kept in memory during sorting steps. The default of 100,000 should be OK for most purposes. If you run into memory problems and cannot increase the memory usage, you can lower this value to 50,000 or even 10,000 (do not enter thousands separators into the box). Even lower values will result in heavy disk usage and poor performance. | |
| − | + | ===Creating an Archive=== | |
| + | DwC archives can only be created for web services that support either ABCD 2.06 or HISPID 5. For all other schemas, the button <small>Create DwC Archive</small> will be disabled. Creating a DwC archive will always create an ABCD archive first, which will then be transformed into DwC. So any existing ABCD archive will be overwritten with an updated version, and any existing DwC archives will be replaced by new versions. | ||
| − | < | + | To start the archiving process, make sure ABCD 2.06 is selected as ''destination schema'' (or HISPID 5, if you're using that) and press <small>Create DwC Archive</small>. This will start the XML archiving process and, once that is successfully finished, the DwC archive transformation. If you've already created an XML archive before, you can skip the XML archiving and convert this file directly by clicking the ''DwC'' link next to the archive name listed under ''Existing Archives''. The XML archiving step and the parameter <small>Paging size</small> are described in detail in the [[Archiving#XML_Archives|XML Archiving section]] above. |
| − | + | During the DwC transformation, the log will display the progress and look similar to this: | |
| − | + | [[File:dwcaArchiveProgressNew.png|border]] | |
| − | < | + | You can cancel the transformation process by clicking the <small>Cancel</small> link in the Status/Messages box. When you do this, BioCASe will try to terminate the process. Depending on the operating system and machine characteristics, this can take some time. Please be patient and wait for the message ''Process terminated'' to appear in the status box. |
| − | == | + | During the process, the first lines of the log output will drop out of view. You can use the <small>Show full log</small> link to open the full log in a separate browser tab. During the conversion process, you can navigate away from the page or close it. When you return and the conversion is still running, you will see the current log output again. If the conversion failed or was cancelled, the log can be viewed from the ''Logs of aborted/failed runs'' box that appears. |
| − | + | ||
| − | XML | + | If the DwC archiving was successful, the line ''SUCCESS: Conversion finished.'' will be printed at the end of the output. The archive(s) generated will be named after the dataset titles and show up under ''Existing archives'', with links for downloading/removing and for re-opening the log file. |
| + | |||
| + | ===Trouble Shooting=== | ||
| + | If you see the line ''ERROR: The DwC Archive creation failed!'', something went wrong. | ||
| + | |||
| + | When trying to find the error, always have a look at the full log file instead of just the few last lines printed in the log output box. Either use the ''Show full log'' link or the ''Show'' link below ''Logs of aborted/failed runs''. Start reading the log from top to bottom and find the first error (a line starting with ''ERROR''). Once you've solved this first problem, don't pay attention to any follow-up errors, since they might be caused by the first problem. Instead, restart the transformation using the ''DwC'' link of the respective XML archive and see if the problem is gone. | ||
| + | |||
| + | Below are some typical error messages: | ||
| + | |||
| + | <syntaxhighlight> | ||
| + | [Errno 13] Permission denied: 'C:\\Workspace\\bps2\\archive\\FloraExsiccataBavarica\\FloraExsiccataBavarica_DwCA_1.0.log' | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | Access is denied for the archiving temp directory. Use the libs test page (section ''Status of writable directories and files'') to make sure Python has full write privileges to this folder. | ||
| + | |||
| + | <syntaxhighlight> | ||
| + | ERROR 05-07 17:06:47,374 - Error trying to remove old status file: [Error 5] Access denied: u'C:\\Workspace\\bps2\\archive\\FloraExsiccataBavarica\\duplicate.txt' | ||
| + | ERROR 05-07 17:06:52,372 - ERROR: The DwC Archive creation failed! | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | Same problem as above, status file ''duplicate.txt'' cannot be removed. | ||
| + | |||
| + | <syntaxhighlight> | ||
| + | INFO 05-07 16:41:14,386 - Starting the transformation engine | ||
| + | INFO 05-07 16:41:14,386 - Java heap space used is 6144M, sort size is 100000 | ||
| + | ERROR 05-07 16:41:14,388 - Java process could not be started: [Error 2] Das System kann die angegebene Datei nicht finden | ||
| + | ERROR 05-07 16:41:14,389 - Check the Test libs page to make sure Java is properly installed and configured. | ||
| + | ERROR 05-07 16:41:19,384 - ERROR: The DwC Archive creation failed! | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | Well - what it reads. | ||
| + | |||
| + | <syntaxhighlight> | ||
| + | INFO 05-07 16:43:42,892 - Starting the transformation engine | ||
| + | INFO 05-07 16:43:42,892 - Java heap space used is 1,024M, sort size is 100000 | ||
| + | Error: Could not create the Java Virtual Machine. | ||
| + | Error: A fatal exception has occurred. Program will exit. | ||
| + | Invalid maximum heap size: -Xmx1,024m | ||
| + | INFO 05-07 16:43:42,908 - Transformation ended with return code 1 | ||
| + | ERROR 05-07 16:43:47,891 - ERROR: The DwC Archive creation failed! | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | Please do not use thousands separators or any unit (MB, GB) for the maximum heap space, just a number: ''1024''. | ||
| + | |||
| + | <syntaxhighlight> | ||
| + | ... | ||
| + | INFO 05-07 16:50:45,359 - Transformation - Loading transformation from XML file [file:///C:/Workspace/bps2/lib/biocase/archive/abcd2.ktr] | ||
| + | INFO 05-07 16:50:45,610 - DwC Conversion - Dispatching started for transformation [DwC Conversion] | ||
| + | INFO 05-07 16:50:45,668 - DwC Conversion - This transformation can be replayed with replay date: 2012/07/05 16:50:45 | ||
| + | INFO 05-07 16:50:45,710 - Get file names - Finished processing (I=0, O=0, R=47, W=47, U=0, E=0) | ||
| + | INFO 05-07 16:50:45,718 - Get parameters - Finished processing (I=0, O=0, R=47, W=329, U=0, E=0) | ||
| + | ERROR 05-07 16:50:55,904 - Sort meta - UnexpectedError: | ||
| + | ERROR 05-07 16:51:16,093 - Sort meta - java.lang.OutOfMemoryError: GC overhead limit exceeded | ||
| + | ... | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | Not enough memory. Go to the System Administration page and increase the memory limit (minimum is 512) or reduce the number of rows kept in memory during sorting. | ||
| + | |||
| + | <syntaxhighlight> | ||
| + | ... | ||
| + | INFO 05-07 16:57:08,435 - Sort occurrences - Finished processing (I=0, O=0, R=2179, W=2179, U=0, E=0) | ||
| + | ERROR 05-07 16:57:08,438 - Abort - Row nr 1 causing abort : [General], [response.1.xml], [file:///C:/Workspace/bps2/archive/FloraExsiccataBavarica/tmp/response.1.xml], [DwC Archive creation], [Unzip file], [null], [2012/07/05 16:57:04.129], [100000], [C:\Workspace\bps2\archive\FloraExsiccataBavarica], [RBG], [Flora exsiccata Bavarica], [1591], [Preserved Specimen], [], [], [], [520], [], [], [], [Germany], [DE], [Puchheim westlich von München; Hausmullschutt auf Moorboden], [], [], [20.08.1915], [], [], [], [], [], [], [], [], [], [], [], [], [], [], [Flora exsiccata Bavarica], [C:\Workspace\bps2\archive\FloraExsiccataBavarica/tmp/Flora_exsiccata_Bavarica occurrence], [], [C:\Workspace\bps2\archive\FloraExsiccataBavarica/duplicate] | ||
| + | ERROR 05-07 16:57:08,438 - Abort - Duplicate catalog number found. | ||
| + | ... | ||
| + | INFO 05-07 16:57:09,762 - Kitchen - Processing ended after 5 seconds. | ||
| + | INFO 05-07 16:57:09,813 - Transformation ended with return code 1 | ||
| + | ERROR 05-07 16:57:09,815 - ERROR: The DwC Archive creation failed! | ||
| + | ERROR 05-07 16:57:09,817 - ERROR: Duplicate catalog number "1591" found in the dataset. Please eliminate it, recreate the XML archive and retry. | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | The ABCD archive contained a duplicate catalog number "1591", which would make the DwC archive invalid. Eliminate that from the database, recreate the XML archive and restart the transformation. | ||
| + | |||
| + | <syntaxhighlight> | ||
| + | ... | ||
| + | INFO 05-07 16:37:01,677 - Removing old files from download folder | ||
| + | ERROR 05-07 16:37:01,679 - [Error 5] Access denied: u'C:\\Workspace\\bps2\\www\\downloads\\EDIT_ATBI\\EDIT_-_ATBI_in_Borjomi_Kharagauli_Georgia.dwca.zip' | ||
| + | ERROR 05-07 16:37:01,680 - ERROR: The DwC Archive creation failed! | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | Access is denied for the download folder. Use the libs test page (section ''Status of writable directories and files'') to make sure Python has full write privileges to this folder. | ||
| + | |||
| + | <syntaxhighlight> | ||
| + | INFO 05-07 17:55:22,793 - DwC Archive creation - Starting entry [Unzip file] | ||
| + | ERROR 05-07 17:57:03,253 - Unzip file - Error trying to process zipped entry [zip:file:///C:/Workspace/bps2/www/downloads/natureinfo/natureinfo_ABCD_2.06.zip!/response.142.xml] from file [file:///C:/Workspace/bps2/www/downloads/natureinfo/natureinfo_ABCD_2.06.zip] ! | ||
| + | ERROR 05-07 17:57:03,253 - Unzip file - java.io.IOException: Not enough space on volume. | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | During the transformation, the XML archive will be unpacked temporarily. This requires some disk space, which wasn't available in this case. | ||
| − | == | + | == Automating Archiving == |
| − | + | Starting with version 3.3 of the BioCASe Provider Software, archiving can be triggered from outside the configuration tool by using a deep link. This allows for scheduling of the archiving process; for example, an ABCD archive could be created once a month to be harvested by GBIF. Both XML and Darwin Core Archive generation can be triggered; moreover, the status of the archiving engine can be retrieved together with the log of the latest archiving process. | |
| − | + | ===Usage and Parameters=== | |
| − | + | Assuming you have a data source called ''pontaurus'' with the access point http://localhost/biocase/pywrapper.cgi?dsa=pontaurus, the URL to be used for triggering archiving would be http://localhost/biocase/archiving.cgi?dsa=pontaurus. | |
| − | |||
| − | |||
| − | + | The following lists the parameters that can be specified. Only ''dsa'' and ''pw'' are mandatory. | |
| − | |||
| − | |||
| − | |||
| − | + | ;dsa (Mandatory): Specifies the data source you want to administer. | |
| − | + | ;pw (Mandatory): Config tool password, either for the whole installation or the data source, if a data source specific password has been set. | |
| − | |||
| − | + | ;action (Optional): This specifies the task to be done. Can be one of the values ''log'', ''cancel'', ''xml'' and ''dwc'', each with the following meaning: | |
| − | + | :{| class="wikitable" | |
| + | |- | ||
| + | ! Value | ||
| + | ! Description | ||
| + | |- | ||
| + | | log | ||
| + | | This is the default action. It returns the status of the archiving engine for the given data source and lists the log for the latest archiving process. | ||
| + | |- | ||
| + | | cancel | ||
| + | | Cancels any running archiving process for the given data source. If no process is running, it will do nothing. | ||
| + | |- | ||
| + | | xml | ||
| + | | Starts XML archiving. You can customize the process with the optional parameters ''schema'' and ''filesize''. | ||
| + | |- | ||
| + | | dwc | ||
| + | | Starts XML archiving with subsequent DarwinCore Archive transformation. You can customize the process with the optional parameters ''schema'' and ''filesize''. Remember that Java needs to be installed for DarwinCore Archiving. | ||
| + | |} | ||
| − | + | ;schema (Optional): Data schema that will be used for storing the published records in the archive. The existing schemas for a data source are listed in the <small>Schemas</small> section on the overview page of the data source configuration. Sample values would be ''ABCD_2.06'' and ''HISPID_5'' (note that the trailing .xml must not be included in this parameter). The default schema that will be used is ABCD 2.06 (identified by schema namespace ''http://www.tdwg.org/schemas/abcd/2.06''); so if that’s what you want, there is no need to specify this parameter. Also no need to specify this parameter if only one schema is mapped; BioCASe will use this schema mapping file. | |
| − | |||
| − | + | :HISPID 5 users: If several schemas are mapped for the data source and the default schema ABCD 2.06 is not used, BioCASe will try to find a mapping file for HISPID 5 (identified by namespace ''http://www.chah.org.au/schemas/hispid/5''); so if you have HISPID 5 mapped, but not ABCD 2.06, you can also omit this parameter and let BioCASe find the mapping file for you. | |
| − | + | ;filesize (Optional): Number of records to be stored in a single file of the archive; default is 1,000. If the number of records per request has been limited for the given data source to a value smaller than this, the file size will be adjusted automatically. This parameter corresponds to the paging size that can be set on the archiving page. | |
| − | |||
| − | |||
| − | + | ===Return values of the script=== | |
| − | + | ;For action ''cancel'': The script will return a message once the process has been cancelled successfully: ''Process cancelled.'' | |
| − | + | ;For action ''log'': The log returned will look similar to this: | |
| − | <pre>. | + | <pre> |
| + | Idle. | ||
| − | + | Below you'll find the log of the latest archiving process. | |
| − | + | ************************************************************************************************************************ | |
| + | INFO 02-10 11:51:49,527 - Starting the transformation engine | ||
| + | INFO 02-10 11:51:49,527 - Java heap space used is 6144M, sort size is 100000 | ||
| + | INFO 02-10 11:51:55,299 - Using "C:\Workspace\bps2\archive\BoBO\tmp\vfs_cache" as temporary files store. | ||
| + | INFO 02-10 11:51:55,439 - Kitchen - Start of run. | ||
| + | INFO 02-10 11:51:55,533 - DwC Archive creation - Start of job execution | ||
| + | (snipped) | ||
| − | = | + | Below you'll find the log of the corresponding XML archiving process. |
| − | + | ************************************************************************************************************************ | |
| + | INFO 02-10 11:51:47,951 - Archive file: C:\Workspace\bps2\archive\BoBO\BoBO_ABCD_2.06.zip | ||
| + | INFO 02-10 11:51:47,951 - Query file: C:\Workspace\bps2\archive\BoBO\BoBO_ABCD_2.06.zip.query.xml | ||
| + | INFO 02-10 11:51:47,951 - Config file: C:\Workspace\bps2\archive\BoBO\BoBO_ABCD_2.06.zip.config.xml | ||
| + | INFO 02-10 11:51:47,951 - Datasource: BoBO | ||
| + | INFO 02-10 11:51:47,951 - Download folder: C:\Workspace\bps2\www\downloads\BoBO | ||
| + | INFO 02-10 11:51:47,951 - Wrapper URL: http://localhost/biocase/pywrapper.cgi?dsa=BoBO | ||
| + | INFO 02-10 11:51:47,951 - Ticket: 80f2963d09b8e9deb6c8f667262df0b9 | ||
| + | INFO 02-10 11:51:47,951 - Limit: 1000 | ||
| + | INFO 02-10 11:51:47,967 - Requesting records 1 - 1000 | ||
| + | INFO 02-10 11:51:48,497 - Hits: 9 | ||
| + | INFO 02-10 11:51:48,497 - Count: 9 | ||
| + | INFO 02-10 11:51:48,497 - Dropped: 0 | ||
| + | INFO 02-10 11:51:48,497 - Status: ok | ||
| + | INFO 02-10 11:51:48,497 - Message: response.1.xml stored in archive BoBO_ABCD_2.06.zip | ||
| + | INFO 02-10 11:51:49,511 - SUCCESS: Archiving finished. | ||
| + | </pre> | ||
| − | + | ;For actions ''xml'' and ''dwc'': If an archiving process has been started successfully, it will not return any result and time out eventually. For these actions, use separate ''log'' requests to find out about the progress of the process. | |
| − | + | ;Possible error messages: The script will return an error message if the requested action cannot be performed. Below you’ll find the possible errors returned: | |
| + | :*''Invalid Datasource given!'' | ||
| + | :*''The specified datasource doesn't have any schemas mapped!'' | ||
| + | :*''The specified schema mapping doesn't exist for this datasource!'' | ||
| + | :*''No password provided!'' | ||
| + | :*''Authentication failed!'' | ||
| + | :*''Unrecognized value for parameter action. Only 'log', 'xml', 'dwc' and 'cancel' are allowed.'' | ||
| + | :*''DarwinCore archives can only be created for the ABCD 2.06 and HISPID 5 schemas, so make sure the datasource supports one of these. If you don't specify a schema in the request, BioCASe will try to use ABCD 2.06, otherwise HIDPID 5.'' | ||
| − | + | ===Examples=== | |
| + | Assuming you have a data source called pontaurus with the access point http://localhost/biocase/pywrapper.cgi?dsa=pontaurus, valid requests would be | ||
| − | + | ;http://localhost/biocase/archiving.cgi?dsa=pontaurus&pw=foo: for returning the status of the archiving engine for data source pontaurus (Idle/Running) and the log for the latest archiving process. | |
| − | + | ;http://localhost/biocase/archiving.cgi?dsa=pontaurus&pw=foo&action=xml: to start XML archiving for any existing ABCD 2.06 mapping with default file size (1000). | |
| − | |||
| − | |||
| − | |||
| − | + | ;http://localhost/biocase/archiving.cgi?dsa=pontaurus&pw=foo&action=xml&filesize=500&schema=HISPID_5: to start XML archiving for HIDPID 5 with filesize=500. | |
| − | === | + | ;http://localhost/biocase/archiving.cgi?dsa=pontaurus&pw=foo&action=dwc: to start XML archiving for any existing ABCD 2.06 mapping; after that has finished, this ABCD archive will be converted into DwC archives. |
| − | |||
| − | + | ;http://localhost/biocase/archiving.cgi?dsa=pontaurus&pw=foo&action=cancel: to cancel any running XML or DwC archiving process. | |
| − | ''' | + | == Discovery of Archives == |
| − | + | Archives available for a given data source can be discovered through the so-called dataset inventory. To get this, just append ''&inventory=1'' to the access URL of the data source, for example http://ww3.bgbm.org/biocase/pywrapper.cgi?dsa=Herbar&inventory=1. | |
| − | + | The dataset inventory response looks roughly like this (diagnostic messages have been removed for brevity): | |
| − | + | <syntaxhighlight lang="XML"> | |
| − | + | <dsi:inventory xmlns:dsi="http://www.biocase.org/schemas/dsi/1.0" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" xsi:schemaLocation="http://www.biocase.org/schemas/dsi/1.0 http://www.bgbm.org/biodivinf/schema/dsi_1_0.xsd"> | |
| + | <!--XML generated by BioCASE PyWrapper software version 3.6.4. Made in Berlin.--> | ||
| + | <dsi:status>OK</dsi:status> | ||
| + | <dsi:created>2017-11-01T10:58:38.109000</dsi:created> | ||
| + | <dsi:service_url>http://ww3.bgbm.org/biocase/pywrapper.cgi?dsa=Herbar</dsi:service_url> | ||
| + | <dsi:datasets> | ||
| + | <dsi:dataset> | ||
| + | <dsi:title>Herbarium Berolinense</dsi:title> | ||
| + | <dsi:id>Herbarium Berolinense</dsi:id> | ||
| + | <dsi:archives> | ||
| + | <dsi:archive filesize="20187103" modified="2017-04-20T12:59:33.810767" namespace="http://www.tdwg.org/schemas/abcd/2.06" rcount="206223">http://ww3.bgbm.org/biocase/downloads/Herbar/Herbarium%20Berolinense.ABCD_2.06.zip</dsi:archive> | ||
| + | <dsi:archive filesize="12373841" modified="2017-04-20T13:16:03.772798" rcount="206223" rowType="http://rs.tdwg.org/dwc/terms/Occurrence">http://ww3.bgbm.org/biocase/downloads/Herbar/Herbarium%20Berolinense.DwCA.zip</dsi:archive> | ||
| + | </dsi:archives> | ||
| + | </dsi:dataset> | ||
| + | </dsi:datasets> | ||
| + | <dsi:diagnostics> | ||
| + | ... | ||
| + | </dsi:diagnostics> | ||
| + | </dsi:inventory> | ||
| − | + | </syntaxhighlight> | |
| − | |||
| − | If | + | The dataset inventory will list all datasets published by a given data source, each with its title and ID. If archives exist for this dataset, they will be listed in ''archives'', each ''archive'' element storing the URL for the respective file. For XML archives, the attribute ''namespace'' will store the namespace of the schema, for DarwinCore archives there will an attribute ''rowType'' with the value <nowiki>http://rs.tdwg.org/dwc/terms/Occurrence</nowiki>. The attribute ''rcount'' specifies the number of records stored in the archive file, ''filesize'' and ''modified'' are hopefully self-explanatory. |
| − | < | ||
| − | + | You can also view the dataset inventory using the manual QueryForm: Scroll down to the section ''Configuration Info'' and click on the link ''Inventory''. After a second, it will appear in the text box below. | |
| − | |||
Latest revision as of 10:04, 1 October 2018
This page describes the archiving features in version 3.4 or later.
If you're using version 3.0 or 3.1, please read the version 3.0/3.1 page, for versions 3.2 and 3.3, go here.
With the archiving feature, you can create archive files that store all data published by your BioCASe web service. That might be handy if your database stores a huge number of records (several hundred thousand or even millions) and the traditional harvesting approach of paging through the records published by the web service takes unacceptably long. Once stored in archives, these files can be downloaded from your web server or sent to the harvester via email and ingested much faster. The OpenUp! natural history aggregator, for example, can use this method for fast harvesting of huge datasets.
There are two types of archives: XML Archives simply store the records as compressed XML documents, each holding a customizable number of records. If your web service publishes 500,000 records by using the ABCD data schema, for example, an ABCD Archive created for that service could hold 500 ABCD documents, each storing 1,000 records. For each schema supported by a BioCASe web service, separate XML archives can be created. You can download a sample ABCD Archive here: File:AlgenEngelsSmall ABCD 2.06.zip.
The BioCASe Provider Software can create XML archives for each schema supported by a web service. For the ABCD 2.06 and HISPID 5 schemas, the XML Archiving is dataset-aware, that means each dataset published by the web service will end up in a separate archive file. All other schemas (including ABCD 1.2) will result in one single file for a given web service.
DarwinCore Archives (DwCAs), in contrast, consist of one or several text files storing the core records and related information, zipped up together with a descriptor file and a metadata document. A detailed specification can be found on the TDWG site, a sample archive here: File:Desmidiaceae Engels.zip.
Information published by web services using the ABCD 2.06 or HISPID 5 schema (dubbed as "ABCD/HISPID dumps") can be stored as DarwinCore Archives. Unsurprisingly, DwCAs make use of the DarwinCore data standard, which is flat and less complex than ABCD 2.06 or HISPID 5. Consequently, a DarwinCore archive will store less information than the corresponding XML archive for a given dataset.
Contents
Managing Archives
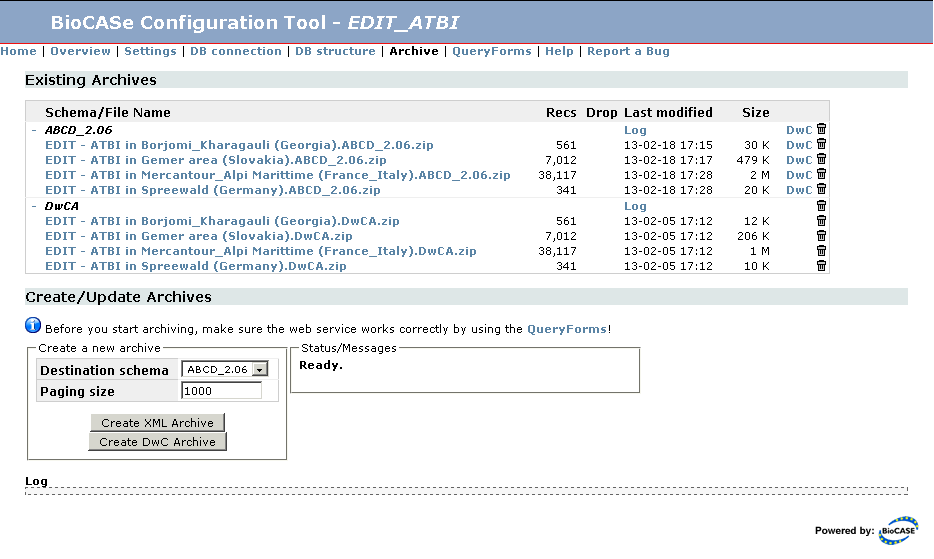
The section Existing Archives allows you download, remove and convert archives or view the log of the archiving process:
The archives are grouped by schema, with DarwinCore archives making up an additional group. You can use the minus or plus symbol next to the group header to expand or collapse all files of that group; for web services publishing dozens of datasets, this might be handy to reduce scrolling on the page. The Log link in the group header will open the log file for the respective archiving process in a separate tab.
For each archive file, the number of records stored in this file, the date of creation and the file size are listed. XML archives may have a number printed in the Dropped column, meaning some records have been dropped due to missing mandatory data elements. This doesn't mean the archive is invalid, but you should review the mapping or your source data to find the reasons for these lost records. Clicking the archive name will download the archive file, so it can be saved to disk or opened with an archive viewing tool. Pressing the trash symbol at the very right will delete the archive; if you use the trash symbol in the group header, all archives of this schema/type will be removed.
For ABCD 2.06 and HISPID 5 XML archives, you will see an additional DwC link next to the trash symbol. This allows you to selectively convert a single dataset from XML to DarwinCore format. In contrast to using the Create DwC Archive button described below, this will spare the XML archiving of the whole web service and just create a DarwinCore archive for the selected dataset, which will be significantly faster. If you use the link in the group header, all archives of the schema will be converted (again without triggering XML archiving first).
XML Archives
If you’ve finished mapping a schema (see ABCD2Mapping to learn how to map ABCD, for example) and tested the resulting web service, you can create XML Archives for that web service. To go to the archiving page, simply use the Archive link on the overview page of the datasource setup. If you haven’t mapped a schema, you won’t be able to create any archives. Before archiving, you should always make sure the web service returns correct results by using the QueryForms.
The section Create/Update Archives allows you to create new XML archive(s) for each schema mapped. Simply select the schema and press Create XML Archive. As mentioned above, archiving ABCD 2.06 or HISPID 5 is dataset-aware, resulting in one XML archive per dataset published. For all other schemas, a single archive file will be created.
The paging size specifies how many records will be bundled into one XML document inside the archive; the default 1,000 should be suitable for most cases. Be aware that a BioCASe web service can restrict the number of records per request - if that limit is below the paging size set for archiving, the paging size will be adjusted automatically after the first packet of records was received from the web service.
During the archiving process, the Log will display messages indicating the progress:
You can always cancel the archiving process by pressing the Cancel link in the Status/Messages box. However, it will take some time for the cancelling to show effect, namely until the current paging step has been completed. Depending upon the paging size set and the speed of your server, this can be up to one minute.
As the archiving progresses, the first lines of the log will drop out of view. Pressing Show full log will open the full log in a separate browser tab. You can navigate away from the archiving page during the process (or simply close it) and return later. Once finished, the dump will show up in the Existing Archives list, or, if it’s still being processed, as Processing in the Status/Messages box. If the process failed for some reason (or you decided to cancel it), a new box Logs of aborted/failed runs will be displayed with a link for displaying the log file. This will allow you to find out what caused the abortion.
For a given web service, you can only create XML archives for one schema at a time. Even though you can start archiving for several web services of your BioCASe installation in parallel, it is not a good idea. The archiving process will put heavy load on your servers – both the database server for data retrieval and the BioCASe server for creating the XML documents. So it is advisable to create just one archive at a time in order to allow both servers to respond to other requests.
DarwinCore Archives
For web services that use the ABCD 2.06 or HISPID 5 schema, all information published by the web service can be stored in DarwinCore Archives. The DarwinCore data standard is less complex than ABCD, so in most cases the DarwinCore Archive will not store all information published by the web service. DarwinCore archives store only one dataset; so as with ABCD 2.06 and HISPID 5 XML archives, DwC-archiving a web service might result in several DarwinCore archives.
DarwinCore archives are not created directly, but from the respective ABCD/HISPID archives. So DwC-archiving a web service will first create ACBD archives, which will be converted into the respective DwC archives subsequently.
Preparation
The conversion of ABCD into DwC is done through the Java-based Kettle library. Therefore, you need to have a Java Runtime Environment (version 1.5 or later) installed. You can check this on the library test page under Optional external binaries:
If you don't have one, you can get it here. Important: If you're running BioCASe on a 64bit machine, make sure to get a 64bit Java, since this will boost performance considerably.
Once Java is installed, go to the System Administration (Start > Config Tool > System Administration) to check and adapt the conversion settings to your environment:
Java binary: If you're sure Java is installed, but the test lib page shows not installed, please provide the full path to the Java binary (for example /usr/local/jdk1.6.0/bin/java). If you have several Java versions installed (for example 32bit and 64bit versions), make sure to point to the correct (preferably 64bit) version.
Max memory usage for Java VM (MB): This is the maximum amount of heap memory Java will be allowed to use (in MB). The default of 1024 will be sufficient for small datasets (up to 100,000 records). For medium-sized datasets (up to 1m records), you should set this value 2048. For large datasets with millions of records, a value of 4096 is recommended. If your server has enough free memory, you should be generous with this limit - larger values speed up the transformation process on most machines. However, you should make sure to leave enough memory for other required applications on your server.
Sort buffer size for transformation (number of rows): This is the number of rows kept in memory during sorting steps. The default of 100,000 should be OK for most purposes. If you run into memory problems and cannot increase the memory usage, you can lower this value to 50,000 or even 10,000 (do not enter thousands separators into the box). Even lower values will result in heavy disk usage and poor performance.
Creating an Archive
DwC archives can only be created for web services that support either ABCD 2.06 or HISPID 5. For all other schemas, the button Create DwC Archive will be disabled. Creating a DwC archive will always create an ABCD archive first, which will then be transformed into DwC. So any existing ABCD archive will be overwritten with an updated version, and any existing DwC archives will be replaced by new versions.
To start the archiving process, make sure ABCD 2.06 is selected as destination schema (or HISPID 5, if you're using that) and press Create DwC Archive. This will start the XML archiving process and, once that is successfully finished, the DwC archive transformation. If you've already created an XML archive before, you can skip the XML archiving and convert this file directly by clicking the DwC link next to the archive name listed under Existing Archives. The XML archiving step and the parameter Paging size are described in detail in the XML Archiving section above.
During the DwC transformation, the log will display the progress and look similar to this:
You can cancel the transformation process by clicking the Cancel link in the Status/Messages box. When you do this, BioCASe will try to terminate the process. Depending on the operating system and machine characteristics, this can take some time. Please be patient and wait for the message Process terminated to appear in the status box.
During the process, the first lines of the log output will drop out of view. You can use the Show full log link to open the full log in a separate browser tab. During the conversion process, you can navigate away from the page or close it. When you return and the conversion is still running, you will see the current log output again. If the conversion failed or was cancelled, the log can be viewed from the Logs of aborted/failed runs box that appears.
If the DwC archiving was successful, the line SUCCESS: Conversion finished. will be printed at the end of the output. The archive(s) generated will be named after the dataset titles and show up under Existing archives, with links for downloading/removing and for re-opening the log file.
Trouble Shooting
If you see the line ERROR: The DwC Archive creation failed!, something went wrong.
When trying to find the error, always have a look at the full log file instead of just the few last lines printed in the log output box. Either use the Show full log link or the Show link below Logs of aborted/failed runs. Start reading the log from top to bottom and find the first error (a line starting with ERROR). Once you've solved this first problem, don't pay attention to any follow-up errors, since they might be caused by the first problem. Instead, restart the transformation using the DwC link of the respective XML archive and see if the problem is gone.
Below are some typical error messages:
[Errno 13] Permission denied: 'C:\\Workspace\\bps2\\archive\\FloraExsiccataBavarica\\FloraExsiccataBavarica_DwCA_1.0.log'Access is denied for the archiving temp directory. Use the libs test page (section Status of writable directories and files) to make sure Python has full write privileges to this folder.
ERROR 05-07 17:06:47,374 - Error trying to remove old status file: [Error 5] Access denied: u'C:\\Workspace\\bps2\\archive\\FloraExsiccataBavarica\\duplicate.txt'
ERROR 05-07 17:06:52,372 - ERROR: The DwC Archive creation failed!Same problem as above, status file duplicate.txt cannot be removed.
INFO 05-07 16:41:14,386 - Starting the transformation engine
INFO 05-07 16:41:14,386 - Java heap space used is 6144M, sort size is 100000
ERROR 05-07 16:41:14,388 - Java process could not be started: [Error 2] Das System kann die angegebene Datei nicht finden
ERROR 05-07 16:41:14,389 - Check the Test libs page to make sure Java is properly installed and configured.
ERROR 05-07 16:41:19,384 - ERROR: The DwC Archive creation failed!Well - what it reads.
INFO 05-07 16:43:42,892 - Starting the transformation engine
INFO 05-07 16:43:42,892 - Java heap space used is 1,024M, sort size is 100000
Error: Could not create the Java Virtual Machine.
Error: A fatal exception has occurred. Program will exit.
Invalid maximum heap size: -Xmx1,024m
INFO 05-07 16:43:42,908 - Transformation ended with return code 1
ERROR 05-07 16:43:47,891 - ERROR: The DwC Archive creation failed!Please do not use thousands separators or any unit (MB, GB) for the maximum heap space, just a number: 1024.
...
INFO 05-07 16:50:45,359 - Transformation - Loading transformation from XML file [file:///C:/Workspace/bps2/lib/biocase/archive/abcd2.ktr]
INFO 05-07 16:50:45,610 - DwC Conversion - Dispatching started for transformation [DwC Conversion]
INFO 05-07 16:50:45,668 - DwC Conversion - This transformation can be replayed with replay date: 2012/07/05 16:50:45
INFO 05-07 16:50:45,710 - Get file names - Finished processing (I=0, O=0, R=47, W=47, U=0, E=0)
INFO 05-07 16:50:45,718 - Get parameters - Finished processing (I=0, O=0, R=47, W=329, U=0, E=0)
ERROR 05-07 16:50:55,904 - Sort meta - UnexpectedError:
ERROR 05-07 16:51:16,093 - Sort meta - java.lang.OutOfMemoryError: GC overhead limit exceeded
...Not enough memory. Go to the System Administration page and increase the memory limit (minimum is 512) or reduce the number of rows kept in memory during sorting.
...
INFO 05-07 16:57:08,435 - Sort occurrences - Finished processing (I=0, O=0, R=2179, W=2179, U=0, E=0)
ERROR 05-07 16:57:08,438 - Abort - Row nr 1 causing abort : [General], [response.1.xml], [file:///C:/Workspace/bps2/archive/FloraExsiccataBavarica/tmp/response.1.xml], [DwC Archive creation], [Unzip file], [null], [2012/07/05 16:57:04.129], [100000], [C:\Workspace\bps2\archive\FloraExsiccataBavarica], [RBG], [Flora exsiccata Bavarica], [1591], [Preserved Specimen], [], [], [], [520], [], [], [], [Germany], [DE], [Puchheim westlich von München; Hausmullschutt auf Moorboden], [], [], [20.08.1915], [], [], [], [], [], [], [], [], [], [], [], [], [], [], [Flora exsiccata Bavarica], [C:\Workspace\bps2\archive\FloraExsiccataBavarica/tmp/Flora_exsiccata_Bavarica occurrence], [], [C:\Workspace\bps2\archive\FloraExsiccataBavarica/duplicate]
ERROR 05-07 16:57:08,438 - Abort - Duplicate catalog number found.
...
INFO 05-07 16:57:09,762 - Kitchen - Processing ended after 5 seconds.
INFO 05-07 16:57:09,813 - Transformation ended with return code 1
ERROR 05-07 16:57:09,815 - ERROR: The DwC Archive creation failed!
ERROR 05-07 16:57:09,817 - ERROR: Duplicate catalog number "1591" found in the dataset. Please eliminate it, recreate the XML archive and retry.The ABCD archive contained a duplicate catalog number "1591", which would make the DwC archive invalid. Eliminate that from the database, recreate the XML archive and restart the transformation.
...
INFO 05-07 16:37:01,677 - Removing old files from download folder
ERROR 05-07 16:37:01,679 - [Error 5] Access denied: u'C:\\Workspace\\bps2\\www\\downloads\\EDIT_ATBI\\EDIT_-_ATBI_in_Borjomi_Kharagauli_Georgia.dwca.zip'
ERROR 05-07 16:37:01,680 - ERROR: The DwC Archive creation failed!Access is denied for the download folder. Use the libs test page (section Status of writable directories and files) to make sure Python has full write privileges to this folder.
INFO 05-07 17:55:22,793 - DwC Archive creation - Starting entry [Unzip file]
ERROR 05-07 17:57:03,253 - Unzip file - Error trying to process zipped entry [zip:file:///C:/Workspace/bps2/www/downloads/natureinfo/natureinfo_ABCD_2.06.zip!/response.142.xml] from file [file:///C:/Workspace/bps2/www/downloads/natureinfo/natureinfo_ABCD_2.06.zip] !
ERROR 05-07 17:57:03,253 - Unzip file - java.io.IOException: Not enough space on volume.During the transformation, the XML archive will be unpacked temporarily. This requires some disk space, which wasn't available in this case.
Automating Archiving
Starting with version 3.3 of the BioCASe Provider Software, archiving can be triggered from outside the configuration tool by using a deep link. This allows for scheduling of the archiving process; for example, an ABCD archive could be created once a month to be harvested by GBIF. Both XML and Darwin Core Archive generation can be triggered; moreover, the status of the archiving engine can be retrieved together with the log of the latest archiving process.
Usage and Parameters
Assuming you have a data source called pontaurus with the access point http://localhost/biocase/pywrapper.cgi?dsa=pontaurus, the URL to be used for triggering archiving would be http://localhost/biocase/archiving.cgi?dsa=pontaurus.
The following lists the parameters that can be specified. Only dsa and pw are mandatory.
- dsa (Mandatory)
- Specifies the data source you want to administer.
- pw (Mandatory)
- Config tool password, either for the whole installation or the data source, if a data source specific password has been set.
- action (Optional)
- This specifies the task to be done. Can be one of the values log, cancel, xml and dwc, each with the following meaning:
Value Description log This is the default action. It returns the status of the archiving engine for the given data source and lists the log for the latest archiving process. cancel Cancels any running archiving process for the given data source. If no process is running, it will do nothing. xml Starts XML archiving. You can customize the process with the optional parameters schema and filesize. dwc Starts XML archiving with subsequent DarwinCore Archive transformation. You can customize the process with the optional parameters schema and filesize. Remember that Java needs to be installed for DarwinCore Archiving.
- schema (Optional)
- Data schema that will be used for storing the published records in the archive. The existing schemas for a data source are listed in the Schemas section on the overview page of the data source configuration. Sample values would be ABCD_2.06 and HISPID_5 (note that the trailing .xml must not be included in this parameter). The default schema that will be used is ABCD 2.06 (identified by schema namespace http://www.tdwg.org/schemas/abcd/2.06); so if that’s what you want, there is no need to specify this parameter. Also no need to specify this parameter if only one schema is mapped; BioCASe will use this schema mapping file.
- HISPID 5 users: If several schemas are mapped for the data source and the default schema ABCD 2.06 is not used, BioCASe will try to find a mapping file for HISPID 5 (identified by namespace http://www.chah.org.au/schemas/hispid/5); so if you have HISPID 5 mapped, but not ABCD 2.06, you can also omit this parameter and let BioCASe find the mapping file for you.
- filesize (Optional)
- Number of records to be stored in a single file of the archive; default is 1,000. If the number of records per request has been limited for the given data source to a value smaller than this, the file size will be adjusted automatically. This parameter corresponds to the paging size that can be set on the archiving page.
Return values of the script
- For action cancel
- The script will return a message once the process has been cancelled successfully: Process cancelled.
- For action log
- The log returned will look similar to this:
Idle. Below you'll find the log of the latest archiving process. ************************************************************************************************************************ INFO 02-10 11:51:49,527 - Starting the transformation engine INFO 02-10 11:51:49,527 - Java heap space used is 6144M, sort size is 100000 INFO 02-10 11:51:55,299 - Using "C:\Workspace\bps2\archive\BoBO\tmp\vfs_cache" as temporary files store. INFO 02-10 11:51:55,439 - Kitchen - Start of run. INFO 02-10 11:51:55,533 - DwC Archive creation - Start of job execution (snipped) Below you'll find the log of the corresponding XML archiving process. ************************************************************************************************************************ INFO 02-10 11:51:47,951 - Archive file: C:\Workspace\bps2\archive\BoBO\BoBO_ABCD_2.06.zip INFO 02-10 11:51:47,951 - Query file: C:\Workspace\bps2\archive\BoBO\BoBO_ABCD_2.06.zip.query.xml INFO 02-10 11:51:47,951 - Config file: C:\Workspace\bps2\archive\BoBO\BoBO_ABCD_2.06.zip.config.xml INFO 02-10 11:51:47,951 - Datasource: BoBO INFO 02-10 11:51:47,951 - Download folder: C:\Workspace\bps2\www\downloads\BoBO INFO 02-10 11:51:47,951 - Wrapper URL: http://localhost/biocase/pywrapper.cgi?dsa=BoBO INFO 02-10 11:51:47,951 - Ticket: 80f2963d09b8e9deb6c8f667262df0b9 INFO 02-10 11:51:47,951 - Limit: 1000 INFO 02-10 11:51:47,967 - Requesting records 1 - 1000 INFO 02-10 11:51:48,497 - Hits: 9 INFO 02-10 11:51:48,497 - Count: 9 INFO 02-10 11:51:48,497 - Dropped: 0 INFO 02-10 11:51:48,497 - Status: ok INFO 02-10 11:51:48,497 - Message: response.1.xml stored in archive BoBO_ABCD_2.06.zip INFO 02-10 11:51:49,511 - SUCCESS: Archiving finished.
- For actions xml and dwc
- If an archiving process has been started successfully, it will not return any result and time out eventually. For these actions, use separate log requests to find out about the progress of the process.
- Possible error messages
- The script will return an error message if the requested action cannot be performed. Below you’ll find the possible errors returned:
- Invalid Datasource given!
- The specified datasource doesn't have any schemas mapped!
- The specified schema mapping doesn't exist for this datasource!
- No password provided!
- Authentication failed!
- Unrecognized value for parameter action. Only 'log', 'xml', 'dwc' and 'cancel' are allowed.
- DarwinCore archives can only be created for the ABCD 2.06 and HISPID 5 schemas, so make sure the datasource supports one of these. If you don't specify a schema in the request, BioCASe will try to use ABCD 2.06, otherwise HIDPID 5.
Examples
Assuming you have a data source called pontaurus with the access point http://localhost/biocase/pywrapper.cgi?dsa=pontaurus, valid requests would be
- http://localhost/biocase/archiving.cgi?dsa=pontaurus&pw=foo
- for returning the status of the archiving engine for data source pontaurus (Idle/Running) and the log for the latest archiving process.
- http://localhost/biocase/archiving.cgi?dsa=pontaurus&pw=foo&action=xml
- to start XML archiving for any existing ABCD 2.06 mapping with default file size (1000).
- http://localhost/biocase/archiving.cgi?dsa=pontaurus&pw=foo&action=xml&filesize=500&schema=HISPID_5
- to start XML archiving for HIDPID 5 with filesize=500.
- http://localhost/biocase/archiving.cgi?dsa=pontaurus&pw=foo&action=dwc
- to start XML archiving for any existing ABCD 2.06 mapping; after that has finished, this ABCD archive will be converted into DwC archives.
- http://localhost/biocase/archiving.cgi?dsa=pontaurus&pw=foo&action=cancel
- to cancel any running XML or DwC archiving process.
Discovery of Archives
Archives available for a given data source can be discovered through the so-called dataset inventory. To get this, just append &inventory=1 to the access URL of the data source, for example http://ww3.bgbm.org/biocase/pywrapper.cgi?dsa=Herbar&inventory=1.
The dataset inventory response looks roughly like this (diagnostic messages have been removed for brevity):
<dsi:inventory xmlns:dsi="http://www.biocase.org/schemas/dsi/1.0" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" xsi:schemaLocation="http://www.biocase.org/schemas/dsi/1.0 http://www.bgbm.org/biodivinf/schema/dsi_1_0.xsd">
<!--XML generated by BioCASE PyWrapper software version 3.6.4. Made in Berlin.-->
<dsi:status>OK</dsi:status>
<dsi:created>2017-11-01T10:58:38.109000</dsi:created>
<dsi:service_url>http://ww3.bgbm.org/biocase/pywrapper.cgi?dsa=Herbar</dsi:service_url>
<dsi:datasets>
<dsi:dataset>
<dsi:title>Herbarium Berolinense</dsi:title>
<dsi:id>Herbarium Berolinense</dsi:id>
<dsi:archives>
<dsi:archive filesize="20187103" modified="2017-04-20T12:59:33.810767" namespace="http://www.tdwg.org/schemas/abcd/2.06" rcount="206223">http://ww3.bgbm.org/biocase/downloads/Herbar/Herbarium%20Berolinense.ABCD_2.06.zip</dsi:archive>
<dsi:archive filesize="12373841" modified="2017-04-20T13:16:03.772798" rcount="206223" rowType="http://rs.tdwg.org/dwc/terms/Occurrence">http://ww3.bgbm.org/biocase/downloads/Herbar/Herbarium%20Berolinense.DwCA.zip</dsi:archive>
</dsi:archives>
</dsi:dataset>
</dsi:datasets>
<dsi:diagnostics>
...
</dsi:diagnostics>
</dsi:inventory>
The dataset inventory will list all datasets published by a given data source, each with its title and ID. If archives exist for this dataset, they will be listed in archives, each archive element storing the URL for the respective file. For XML archives, the attribute namespace will store the namespace of the schema, for DarwinCore archives there will an attribute rowType with the value http://rs.tdwg.org/dwc/terms/Occurrence. The attribute rcount specifies the number of records stored in the archive file, filesize and modified are hopefully self-explanatory.
You can also view the dataset inventory using the manual QueryForm: Scroll down to the section Configuration Info and click on the link Inventory. After a second, it will appear in the text box below.