Preparation: Difference between revisions
| (13 intermediate revisions by the same user not shown) | |||
| Line 8: | Line 8: | ||
== Metadata == | == Metadata == | ||
Metadata | Metadata describe your dataset as a whole - title, description, curator’s name, ownership, date of last modification, terms of use etc. BioCASe can publish a lot of metadata – and you should take the time to provide as much information as you can, since this will increase the usability of your published data. | ||
For storing metadata, please create an extra table in your database and fill it in. If you go to the BioCASe folder of your installation, you’ll find the template for this table in sampledb | For storing metadata, please create an extra table in your database and fill it in. If you go to the BioCASe folder of your installation, you’ll find the template for this table in ''sampledb/abcdmetadata''. Either use the SQL script to create it in your database, or import it from the Access database (''metadata.mdb''). | ||
Per default, all records of your database will make up one dataset, being described by one set of metadata. In case your database holds records of several datasets, you can also group these into several datasets in ABCD by providing different sets of metadata. So when you realize during gathering of meta-information that they’re not the same for all records, this is an indicator that your database must be grouped into several datasets. | Per default, all records of your database will make up one dataset, being described by one set of metadata. In case your database holds records of several datasets, you can also group these into several datasets in ABCD by providing different sets of metadata. So when you realize during gathering of meta-information that they’re not the same for all records, this is an indicator that your database must be grouped into several datasets. | ||
| Line 19: | Line 19: | ||
As you will see during configuration, the Provider Software is able to join together the different tables of your database’s data model in order to retrieve all information to be published from the database. This works good if the number of tables is within reasonable limits, that is up to approximately 10-15. | As you will see during configuration, the Provider Software is able to join together the different tables of your database’s data model in order to retrieve all information to be published from the database. This works good if the number of tables is within reasonable limits, that is up to approximately 10-15. | ||
Data models used by off-the- | Data models used by off-the-shelf collection management system are usually highly normalised and use far more tables. Specify’s data model, for example, has about 70 tables. Configuring such a complex model in BioCASe will be a terrible pain; moreover, in case your database holds a huge number of records, the time required by the Provider Software for answering search request will become too long. | ||
Therefore it is advisable in such cases to add some controlled denormalisation to your database, that is to export the data to be published into several cache tables. These tables could be stored in the same database as the source tables or separate in an extra cache database, maybe even on a different server. They could be created in a regular interval (e.g. each night) or upon request by SQL scripts. | Therefore it is advisable in such cases to add some controlled denormalisation to your database, that is to export the data to be published into several cache tables. These tables could be stored in the same database as the source tables or separate in an extra cache database, maybe even on a different server. They could be created in a regular interval (e.g. each night) or upon request by SQL scripts. | ||
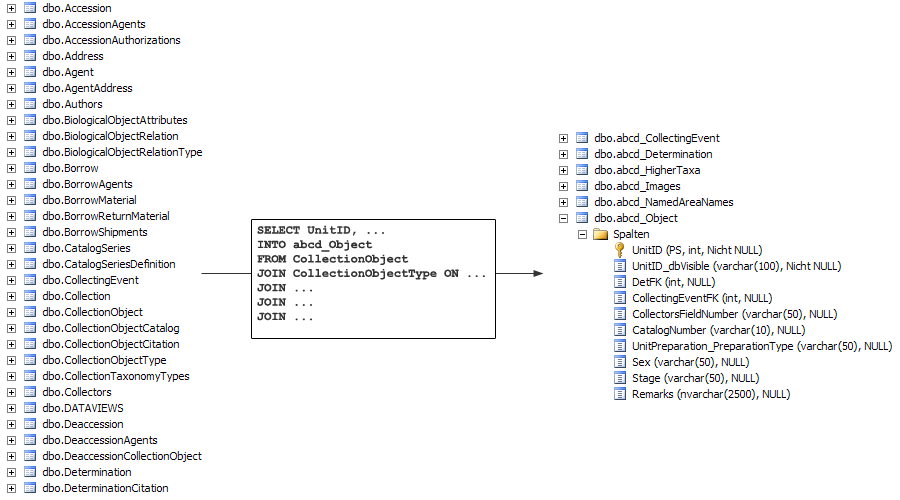
The figure below shows this denormalisation process for the data model of Specify. On the left side, you see some of Specify’s source tables (only the ones starting with the letters A, B, C and D). With an SQL script | The figure below shows this denormalisation process for the data model of Specify. On the left side, you see some of Specify’s source tables (only the ones starting with the letters A, B, C and D). With an SQL script they will be reduced to 7 cache tables, one of them being abcd_object. These tables hold only the columns that will be published, drawn from up to 15 source tables. | ||
[[image:prepDenormalisation.png|750px|border]] | |||
== Repeatable Elements in ABCD == | == Repeatable Elements in ABCD == | ||
One of the main features of ABCD (the schema you will probably use to publish your data) is repeatable elements. For example, ABCD can store multiple identifications or several URLs for multimedia objects attached to a specimen record. All these URLs will be stored in the same ABCD element – namely MultimediaObject/FileURI. | One of the main features of ABCD (the schema you will probably use to publish your data) is repeatable elements. For example, ABCD can store multiple identifications or several URLs for multimedia objects attached to a specimen record. All these URLs will be stored in the same ABCD element – namely ''MultimediaObject/FileURI''. | ||
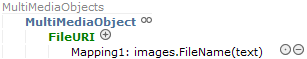
You can tell repeatable elements in ABCD by the existence of a container element. For ''MultimediaObject'', for example, there is a container element ''MultimediaObjects'' one level above which will hold the (potentially) several multimedia objects: | |||
[[File:prepMultimediaObjects.png]] | |||
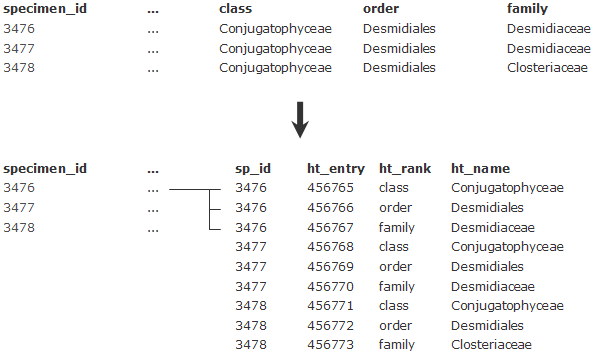
Another example is the higher taxonomy (all taxa above genus level). Typically these are stored in different columns in databases (see figure below). In ABCD, they will all end up in the same element, ''HigherTaxon/Name''. In order to be able to map the different ranks stored in columns to the same ABCD concept, we need to pivot these columns into rows – each rank kept in a column needs to be turned into a row, with two columns storing taxon name and rank, one column for linking to the according specimen record, plus one newly created primary key: | |||
[[File:prepPivoting.png|border]] | |||
Such a transformation can be easily done with a database view. Views do exactly what the name suggests – they provide a different view on the data. They can be used like regular tables, but they merely represent a different representation of another table. Updates of the underlying tables (or insert/deletions) will also affect views created on these tables, since the data is stored only once in the database. | Such a transformation can be easily done with a database view. Views do exactly what the name suggests – they provide a different view on the data. They can be used like regular tables, but they merely represent a different representation of another table. Updates of the underlying tables (or insert/deletions) will also affect views created on these tables, since the data is stored only once in the database. | ||
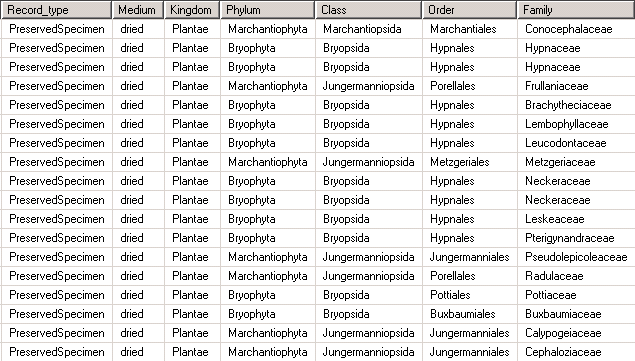
Let’s assume you have the higher taxonomy stored in the specimen table like this: | Let’s assume you have the higher taxonomy stored in the specimen table like this: | ||
[[File:prepEditTable.png]] | |||
The SQL statement | The SQL statement | ||
CREATE VIEW [dbo].[vwHigherTaxa] | <pre> | ||
CREATE VIEW [dbo].[vwHigherTaxa] AS | |||
SELECT 'k_' + [EDIT_ATBI_RecordID] AS id, [EDIT_ATBI_RecordID] AS unit_id, [kingdom] AS name, 'kingdom' AS rank | |||
SELECT 'k_' + [EDIT_ATBI_RecordID] AS id, [EDIT_ATBI_RecordID] AS unit_id, | |||
[kingdom] AS name, 'kingdom' AS rank | |||
FROM unit_data | FROM unit_data | ||
WHERE [kingdom] IS NOT NULL | WHERE [kingdom] IS NOT NULL | ||
UNION | |||
SELECT 'p_' + [EDIT_ATBI_RecordID], [EDIT_ATBI_RecordID], [phylum], ' | UNION ALL | ||
SELECT 'p_' + [EDIT_ATBI_RecordID], [EDIT_ATBI_RecordID], [phylum], 'phylum'‚ | |||
FROM unit_data | FROM unit_data | ||
WHERE [phylum] IS NOT NULL | WHERE [phylum] IS NOT NULL | ||
UNION | |||
UNION ALL | |||
... | ... | ||
</pre> | |||
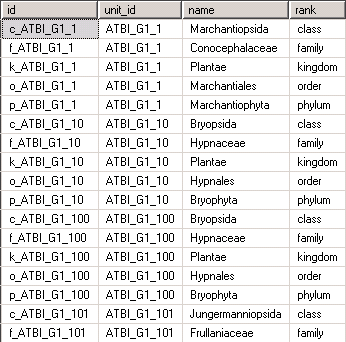
will create a view that meets the requirements described above. Remember that even though it looks like a table, it’s just another representation of the data stored in the table unit_data: | |||
[[File:prepEditView.png]] | |||
Such pivoting must be done in all cases when several columns need to be mapped to the same (repeatable) ABCD element. If it’s just one value to be mapped, there is no need for that. Commonly used ABCD elements that are repeatable and might ask for such preparatory transformations are | Such pivoting must be done in all cases when several columns need to be mapped to the same (repeatable) ABCD element. If it’s just one value to be mapped, there is no need for that. Commonly used ABCD elements that are repeatable and might ask for such preparatory transformations are | ||
If you have problems preparing your database for publication, please contact the | * Identification | ||
* HigherTaxon | |||
* GatheringSite/NamedArea | |||
* Metadata/Scope/GeoecologicalTerms | |||
* Metadata/Scope/TaxonomicTerms | |||
* MultimediaObjects | |||
* MeasurementsOrFacts | |||
If you have problems preparing your database for publication, please [[Special:Contact|contact the BioCASe team]]. | |||
Latest revision as of 10:30, 5 April 2012
Before you start publishing your database with BioCASe, you need to consider several issues. In some cases, some preparation of your database for the publication might be necessary. You should read this page of the wiki careful before you start configuring the Provider Software for a given database.
Live-DB versus Snapshot
The easiest way is to connect BioCASe to the same database you’re working with. This would result in any changes you do being visible on the BioCASe access point right away – which is an idea you might not like. Some curators prefer to publish defined versions of their database, for example after they’ve finished reviewing all specimens of a certain genus. If you prefer this, you need to create a second database that holds defined snapshots of your working database and that will be published instead. Snapshots could be generated in a regular interval (e.g., each Friday evening) or upon request. Please contact your IT guys for more help on this.
Access Control
The BioCASe Provider Software will only publish your data to biodiversity networks, meaning it will only read from the database and never do any changes. For security reasons it’s a good idea to create a user for BioCASe that has only read privileges on the database – more precisely on the tables you want to publish. Since the configuration tool will try to load a list of tables and columns from the database to allow easier setup, this user should also be allowed to load meta-information (SHOW TABLES, SHOW COLUMNS).
Metadata
Metadata describe your dataset as a whole - title, description, curator’s name, ownership, date of last modification, terms of use etc. BioCASe can publish a lot of metadata – and you should take the time to provide as much information as you can, since this will increase the usability of your published data.
For storing metadata, please create an extra table in your database and fill it in. If you go to the BioCASe folder of your installation, you’ll find the template for this table in sampledb/abcdmetadata. Either use the SQL script to create it in your database, or import it from the Access database (metadata.mdb).
Per default, all records of your database will make up one dataset, being described by one set of metadata. In case your database holds records of several datasets, you can also group these into several datasets in ABCD by providing different sets of metadata. So when you realize during gathering of meta-information that they’re not the same for all records, this is an indicator that your database must be grouped into several datasets.
If all records to be published share the same metadata, the metadata table needs to have just one row. If they don’t, it needs to hold several rows, one set of metadata for each dataset, identified by a unique primary key (1, 2, …). In order to associate each record of your database with the correct set of metadata, you will need to add a foreign key into the base record table that links to the respective entry in the metadata table. Ask your IT guys if you need help on that.
Controlled Denormalisation
As you will see during configuration, the Provider Software is able to join together the different tables of your database’s data model in order to retrieve all information to be published from the database. This works good if the number of tables is within reasonable limits, that is up to approximately 10-15.
Data models used by off-the-shelf collection management system are usually highly normalised and use far more tables. Specify’s data model, for example, has about 70 tables. Configuring such a complex model in BioCASe will be a terrible pain; moreover, in case your database holds a huge number of records, the time required by the Provider Software for answering search request will become too long.
Therefore it is advisable in such cases to add some controlled denormalisation to your database, that is to export the data to be published into several cache tables. These tables could be stored in the same database as the source tables or separate in an extra cache database, maybe even on a different server. They could be created in a regular interval (e.g. each night) or upon request by SQL scripts.
The figure below shows this denormalisation process for the data model of Specify. On the left side, you see some of Specify’s source tables (only the ones starting with the letters A, B, C and D). With an SQL script they will be reduced to 7 cache tables, one of them being abcd_object. These tables hold only the columns that will be published, drawn from up to 15 source tables.
Repeatable Elements in ABCD
One of the main features of ABCD (the schema you will probably use to publish your data) is repeatable elements. For example, ABCD can store multiple identifications or several URLs for multimedia objects attached to a specimen record. All these URLs will be stored in the same ABCD element – namely MultimediaObject/FileURI.
You can tell repeatable elements in ABCD by the existence of a container element. For MultimediaObject, for example, there is a container element MultimediaObjects one level above which will hold the (potentially) several multimedia objects:
Another example is the higher taxonomy (all taxa above genus level). Typically these are stored in different columns in databases (see figure below). In ABCD, they will all end up in the same element, HigherTaxon/Name. In order to be able to map the different ranks stored in columns to the same ABCD concept, we need to pivot these columns into rows – each rank kept in a column needs to be turned into a row, with two columns storing taxon name and rank, one column for linking to the according specimen record, plus one newly created primary key:
Such a transformation can be easily done with a database view. Views do exactly what the name suggests – they provide a different view on the data. They can be used like regular tables, but they merely represent a different representation of another table. Updates of the underlying tables (or insert/deletions) will also affect views created on these tables, since the data is stored only once in the database.
Let’s assume you have the higher taxonomy stored in the specimen table like this:
The SQL statement
CREATE VIEW [dbo].[vwHigherTaxa] AS
SELECT 'k_' + [EDIT_ATBI_RecordID] AS id, [EDIT_ATBI_RecordID] AS unit_id,
[kingdom] AS name, 'kingdom' AS rank
FROM unit_data
WHERE [kingdom] IS NOT NULL
UNION ALL
SELECT 'p_' + [EDIT_ATBI_RecordID], [EDIT_ATBI_RecordID], [phylum], 'phylum'‚
FROM unit_data
WHERE [phylum] IS NOT NULL
UNION ALL
...
will create a view that meets the requirements described above. Remember that even though it looks like a table, it’s just another representation of the data stored in the table unit_data:
Such pivoting must be done in all cases when several columns need to be mapped to the same (repeatable) ABCD element. If it’s just one value to be mapped, there is no need for that. Commonly used ABCD elements that are repeatable and might ask for such preparatory transformations are
- Identification
- HigherTaxon
- GatheringSite/NamedArea
- Metadata/Scope/GeoecologicalTerms
- Metadata/Scope/TaxonomicTerms
- MultimediaObjects
- MeasurementsOrFacts
If you have problems preparing your database for publication, please contact the BioCASe team.